Research Article
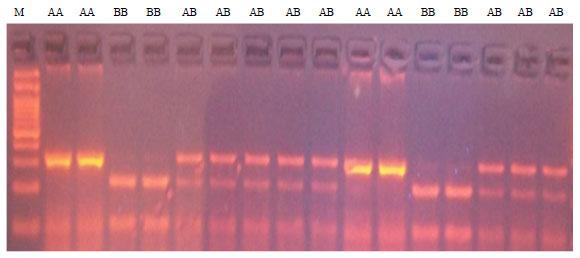
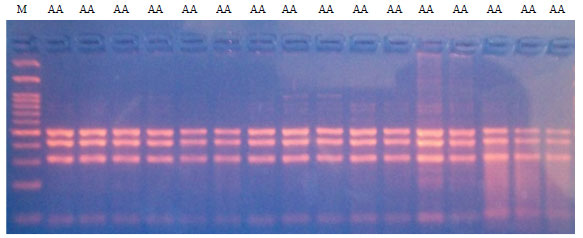
Effect of Genetic Polymorphisms in GH/HpaII and MSTN/DraI Loci on Body Weight in Friesian Bull Calves
Department of Animal Husbandry and Animal Wealth Development, Faculty of Veterinary Medicine, Damanhour University, Egypt
LiveDNA: 20.12509
A. I. Ateya
Department of Animal Husbandry and Animal Wealth Development, Faculty of Veterinary Medicine, Mansoura University, Egypt
LiveDNA: 20.16146
K. M. Sadek
Department of Biochemistry, Faculty of Veterinary Medicine, Damanhour University, Egypt
LiveDNA: 20.15058
H. A. Radwan
Department of Animal Husbandry and Animal Wealth Development, Faculty of Veterinary Medicine, Mansoura University, Egypt