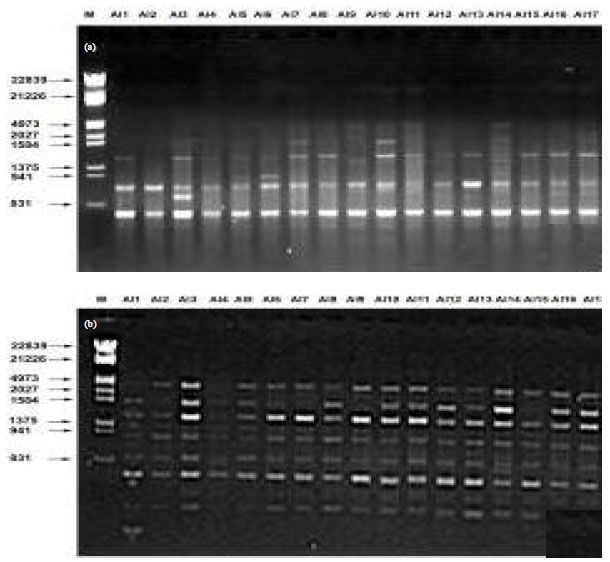
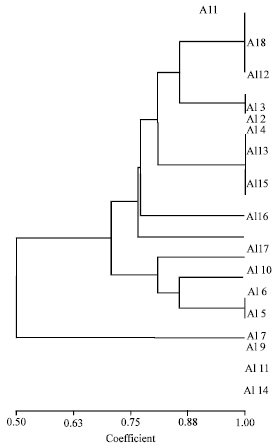
Research Article
Frequency Distribution and Assessment of Genetic Diversity of Novel Endophyte Alternaria alternata Accessions Isolated from Pongamia pinnata L.
Microbial Biotechnology Laboratory, Jaipur National University, Jaipur, India-302017, India