Research Article
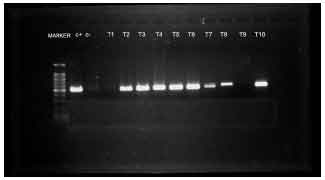
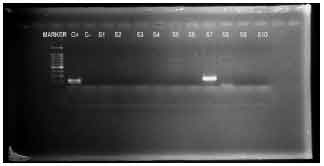
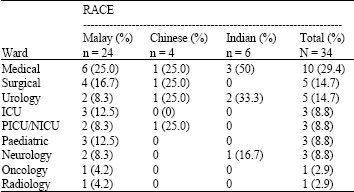
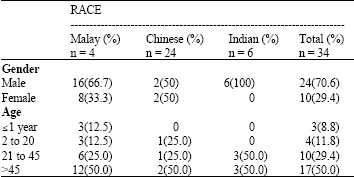
Detection of Betalactamase Producing Bacterial Genes and their Clinical Features
Department of Medical Microbiology and Immunology, Faculty of Medicine, National University Malaysia, Cheras-56000, Kuala Lumpur, Malaysia
S. Husin
Department of Medical Microbiology and Immunology, Faculty of Medicine, National University Malaysia, Cheras-56000, Kuala Lumpur, Malaysia
M. M. Rahman
Department of Medical Microbiology and Immunology, Faculty of Medicine, National University Malaysia, Cheras-56000, Kuala Lumpur, Malaysia