Research Article
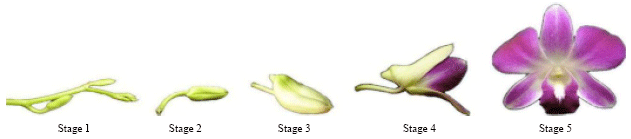
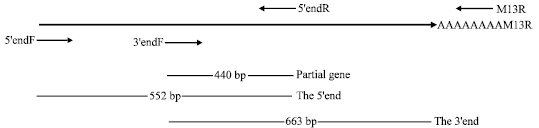
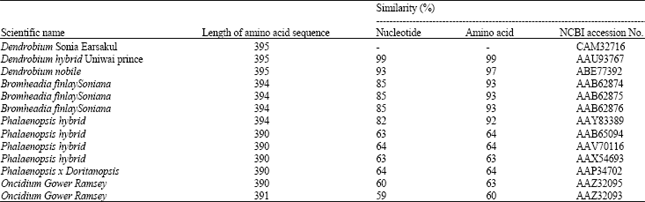
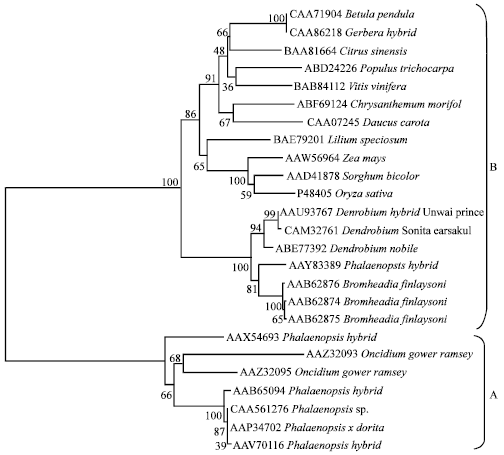
Isolation and Characterization of Chalcone Synthase Gene Isolated from Dendrobium Sonia Earsakul
Center for Agricultural Biotechnology (CAB), Kasetsart University, Kamphang Saen Campus, Nakhon Pathom 73140, Thailand
T. Sutabutra
Department of Plant Pathology, Faculty of Agriculture, Kasetsart University, Bangkok 10900, Thailand
P. Chiemsombat
Department of Plant Pathology, Faculty of Agriculture, Kasetsart University, Kamphang Saen Campus, Nakhon Pathom 73140, Thailand
C. Pitaksutheepong
National Center for Genetic Engineering and Biotechnology (BIOTEC), 113 Phaholyothin Road, Klong 1, Klong Luang, Pathumthani 12120, Thailand