Research Article
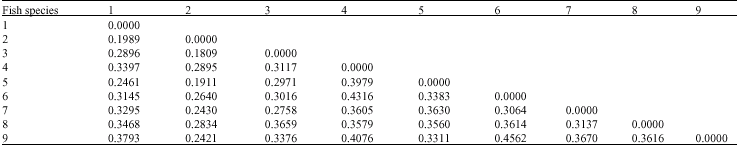
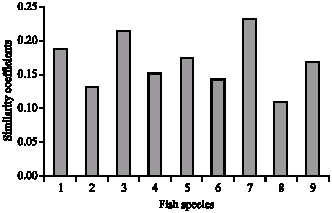
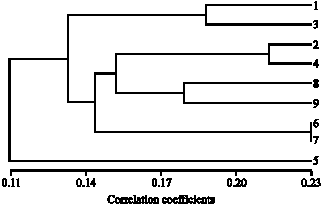
An Identification in Fish of the Genus Puntius Hamilton 1822 (Cypriniformes:Cyprinidae) of Some Wetlands in Northeast Thailand With the Use of Random Amplified Polymorphic DNA Technique
Department of Fisheries, Faculty of Agriculture, Khon Kaen University, Khon Kaen 40002, Thailand
R. Rapley
Department of Biosciences, School of Life Sciences, University of Hertfordshire, College Lane, Hatfield, Herts AL10 9AB, UK
M. Duangjinda
Department of Animal Science, Faculty of Agriculture, Khon Kaen University, Khon Kaen 40002, Thailand
A. Suksri
Faculty of Agriculture, Khon Kaen University, Khon Kaen 40002, Thailand