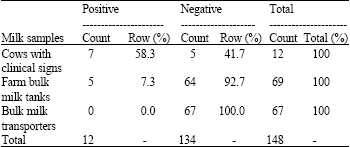
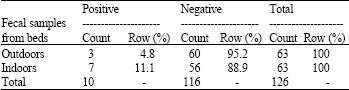Research Article
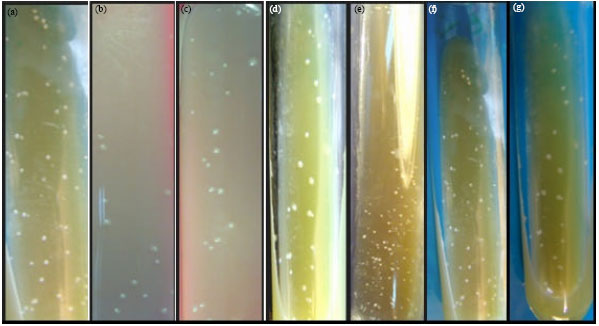
Isolation and Identification of Mycobacterium avium Subsp. paratuberculosis from Milk, Manure and Fecal Samples of Holstein-Friesian Dairy Cattle
Department of Veterinary Research and Biotechnology, Razi Vaccine and Serum Research Institute, Mashhad, Iran
T.Z. Salehi
2Departemnet of Microbiology, Faculty of Veterinary Medicine, University of Tehran, Tehran, Iran
H. Tadjbakhsh
2Departemnet of Microbiology, Faculty of Veterinary Medicine, University of Tehran, Tehran, Iran
M.F. Najafi
Department of Veterinary Research and Biotechnology, Razi Vaccine and Serum Research Institute, Mashhad, Iran
M. Rabbani
Departemnet of Microbiology, Faculty of Veterinary Medicine, University of Tehran, Tehran, Iran