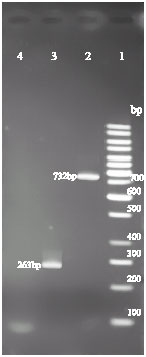
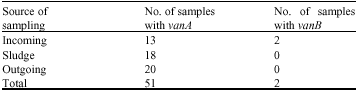Research Article
Vancomycin-Resistant Enterococcus faecalis from a Wastewater Treatment Plant in Tabriz, Iran
Department of Community Dentistry, Tabriz University of Medical Science, Tabriz, Iran
R. Abashov
Department of Microbiology, Baku State University, Baku, Azerbaijan
M. Aghazadeh
Department of Microbiology, Tabriz University of Medical Science, Tabriz, Iran
H. Eslami
Department of Community Dentistry, Tabriz University of Medical Science, Tabriz, Iran
S.G. Oskouei
Department of Community Dentistry, Tabriz University of Medical Science, Tabriz, Iran
D. Mohammadnejad
EM Lab Drug Applied Research Center and Development Complex, Tabriz University of Medical Sciences, Tabriz, Iran