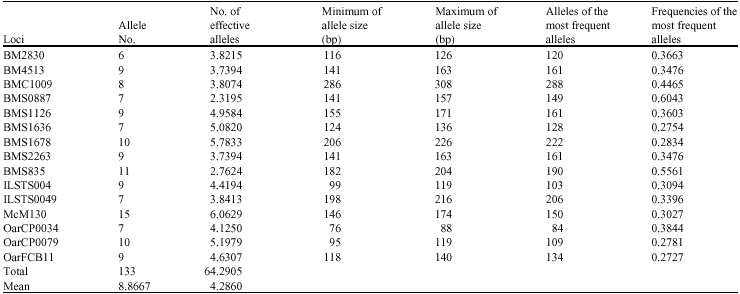
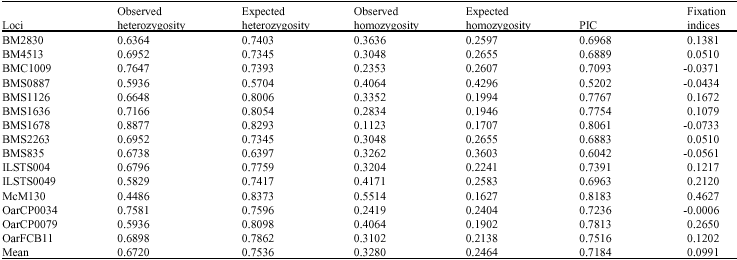
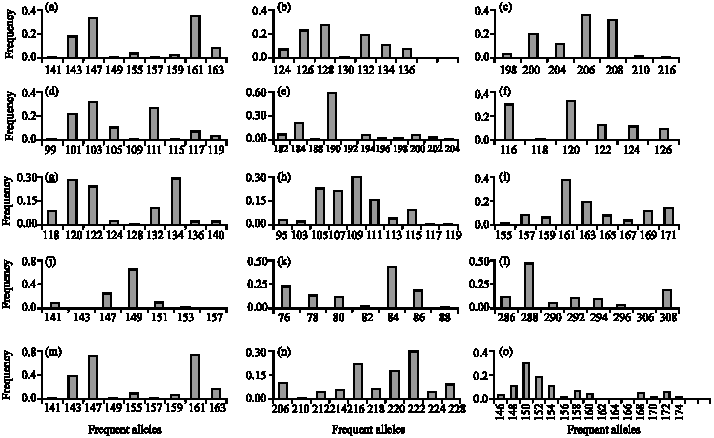
Research Article
Study on Population Genetic Structure of Liangshan Semi-Wool Sheep Using Microsatellite Markers
Department of Animal Science and Technology, Sichuan Agricultural University, Ya`an, China
M.L. Zhou
Department of Animal Science and Technology, Sichuan Agricultural University, Ya`an, China
X.H. Zhang
Department of Life Sciences and Technology, Xinxiang Medical University, Xinxiang, China
D.J. Wu
Department of Animal Science and Technology, Sichuan Agricultural University, Ya`an, China