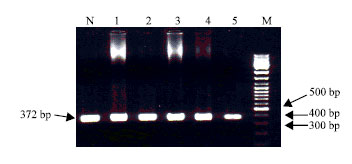
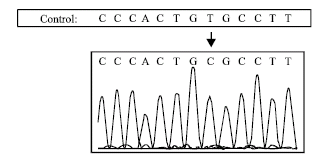
Research Article
Identification of a Novel Missense Mutation in Exon 4 of the Human Factor VIII Gene Associated with Sever Hemophilia a Patient
Islamic Azad University, Science and Research Campus, Tehran, Iran
Mohammad Ali Hosseinpour
Department of Biology, Faculty of Sciences, University of Tabriz, Tabriz, Iran
Sheideh Montaser-Kouhsari
Department of Biology, Faculty of Sciences, University of Tehran, Tehran, Iran
Mohammad Asgharzadeh
Hematology and Oncology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Abbas Ali Hosseinpour
Tabriz University of Medical Sciences, Tabriz, Iran