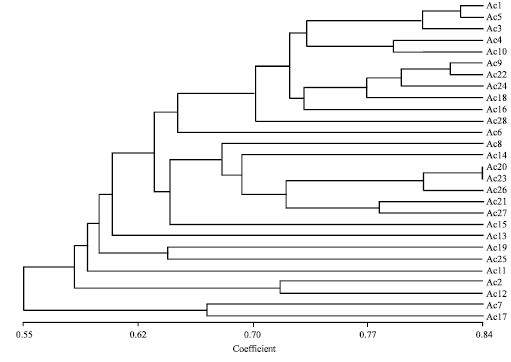
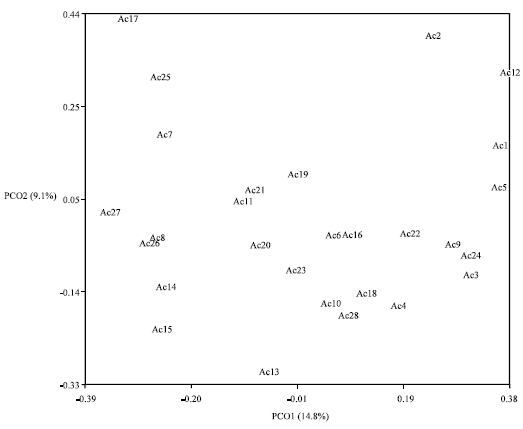
Research Article
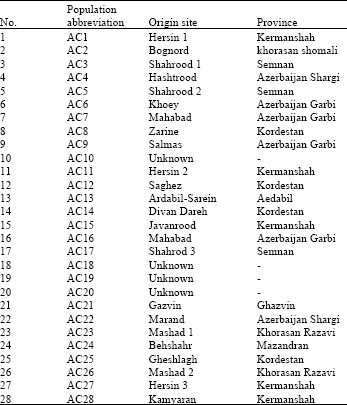
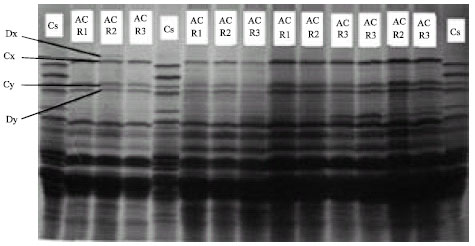
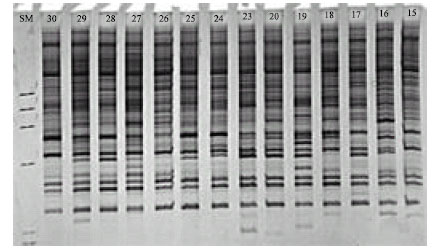
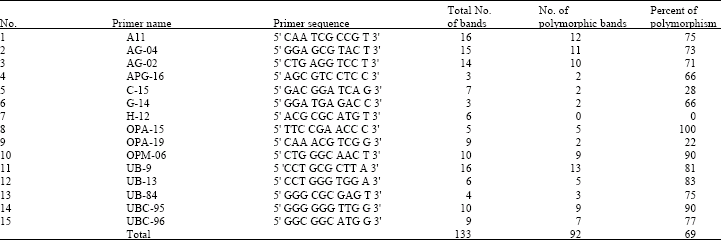
Genetic Variation of Jointed Goatgrass (Aegilops cylindrica Host.) From Iran Using RAPD-PCR and SDS-PAGE of Seed Proteins
Department of Agronomy and Plant Breeding, Agricultural College, University of Tehran, Karaj, Iran
M. R. Naghavi
Department of Agronomy and Plant Breeding, Agricultural College, University of Tehran, Karaj, Iran
S. A. Pyghambari
Department of Agronomy and Plant Breeding, Agricultural College, University of Tehran, Karaj, Iran
Sabokdast
Department of Agronomy and Plant Breeding, Agricultural College, University of Tehran, Karaj, Iran