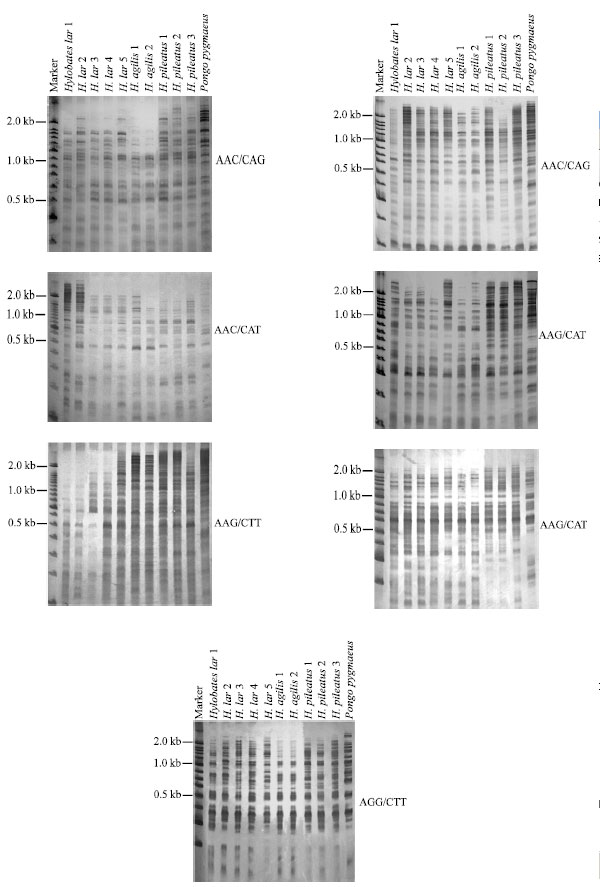
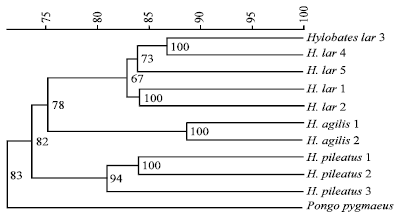
Research Article
Analysis of the Genetic Relationships among Thai Gibbon Species Using AFLP Markers
Department of Biochemistry, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand
Arunrat Chaveerach
Department of Biology, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand
Nison Sattayasai
Department of Biochemistry, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand
Alongkoad Tanomtong
Department of Biology, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand
Scott A. Suarez
Department of Anthropology, Miami University, USA
Suporn Nuchadomrong
Department of Biochemistry, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand