Research Article
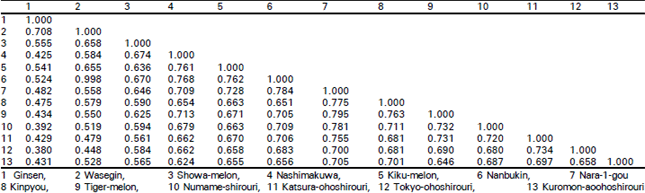
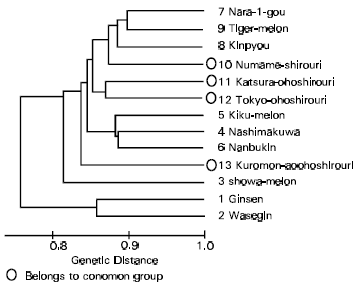
Classification of Oriental Melon by RAPD Analysis
Department of Bioresource Production, Faculty of Agriculture, Kagawa University, Miki-Cho, Kagawa, 761-0795, Japan
Yusuke Kosugi
Department of Bioresource Production, Faculty of Agriculture, Kagawa University, Miki-Cho, Kagawa, 761-0795, Japan
Tomohiro Yanagi
Department of Bioresource Production, Faculty of Agriculture, Kagawa University, Miki-Cho, Kagawa, 761-0795, Japan
Haruo Suzuki
Department of Bioresource Production, Faculty of Agriculture, Kagawa University, Miki-Cho, Kagawa, 761-0795, Japan
Pankaj Kumar Bhowmik
Department of Bioresource Production, Faculty of Agriculture, Kagawa University, Miki-Cho, Kagawa, 761-0795, Japan
Sutevee Sukarakarn
Tropical Vegetable Research Center Kasetsart University, Bangkok 10900, Thailand