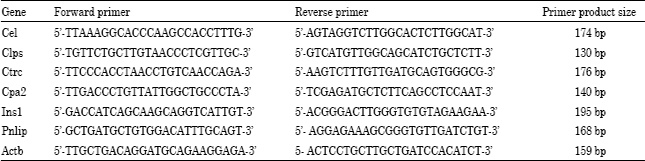
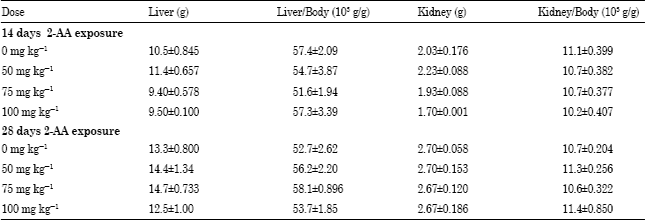
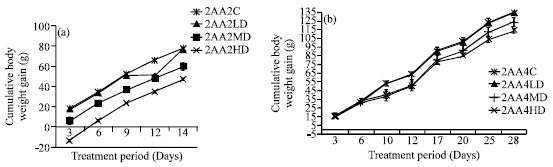
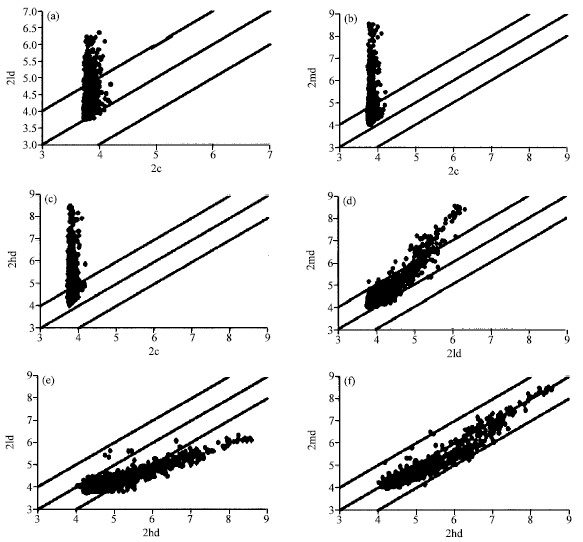
Research Article
Pancreatic Gene Expression Altered Following Dietary Exposure to 2-Aminoanthracene: Links to Diabetogenic Activity
Department of Chemistry and Biochemistry, Southern Illinois University, IL 62901, Carbondale
J.C. Means
Department of Chemistry and Biochemistry, Southern Illinois University, IL 62901, Carbondale