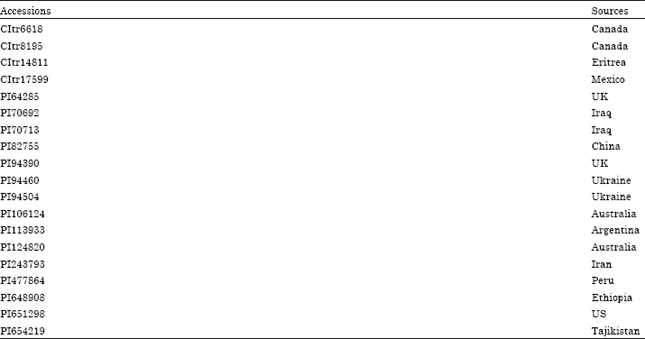
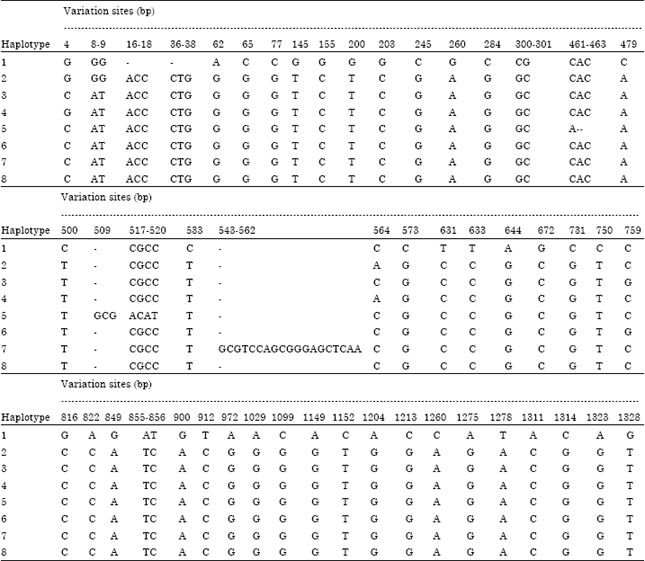
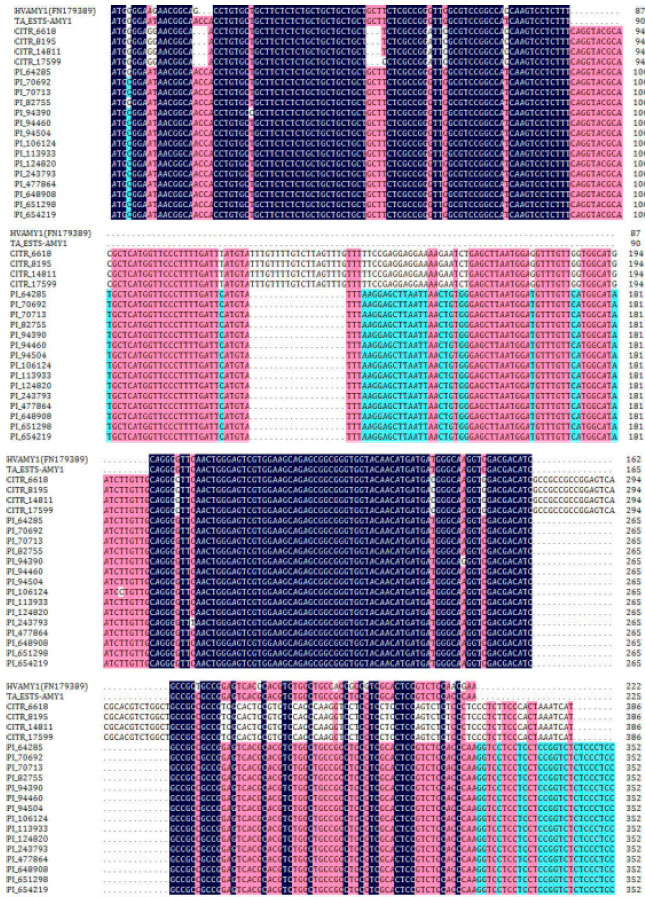
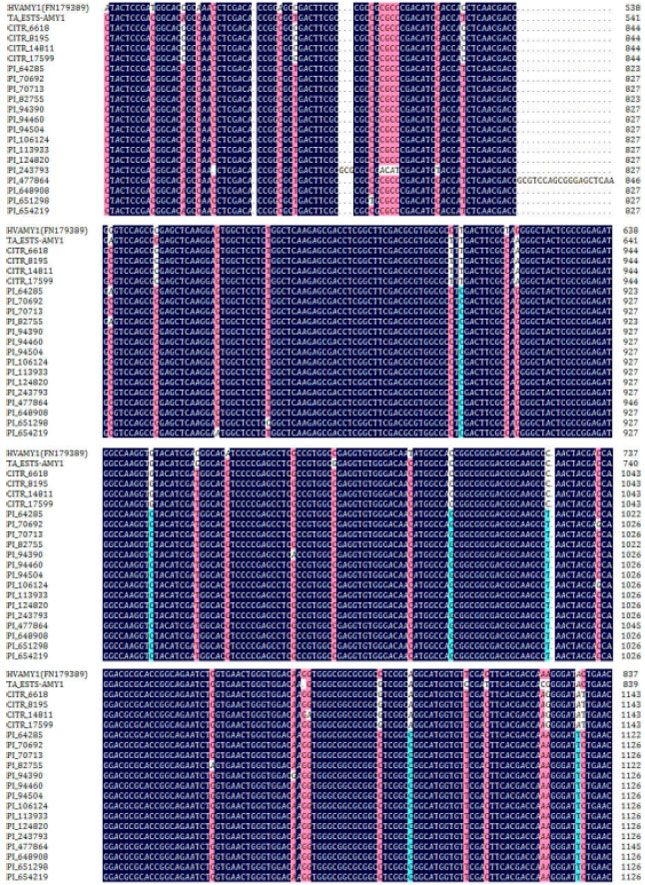
Research Article
Molecular Characterization of the Amy1 Gene in Hexaploid Wheat
Triticeae Research Institute, Sichuan Agricultural University, Wenjiang, Sichuan 611130, China
Yong Zhou
Triticeae Research Institute, Sichuan Agricultural University, Wenjiang, Sichuan 611130, China
Jia Li
Triticeae Research Institute, Sichuan Agricultural University, Wenjiang, Sichuan 611130, China
Yu-Jiao Liu
Triticeae Research Institute, Sichuan Agricultural University, Wenjiang, Sichuan 611130, China
Ya-Xi Liu
Triticeae Research Institute, Sichuan Agricultural University, Wenjiang, Sichuan 611130, China