Research Article
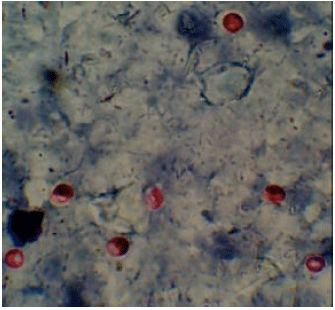
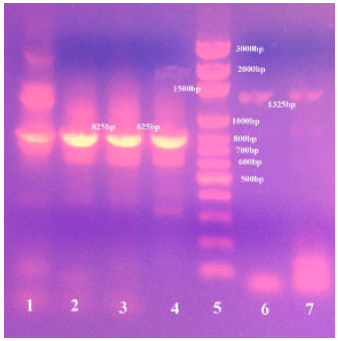
Prevalence and Molecular Discrimination of Cryptosporidium parvum in Calves in Behira Provinces, Egypt
Department of Zoonosis, Veterinary Research Division, National Research Center, Dokki, Giza, Egypt
Fathia A.M. Khalil
Department of Parasitology and Animal Diseases, National Research Center, Dokki, Giza, Egypt
K.A. AbdEl-Razik
Department of Animal Reproduction, National Research Center, Dokki, Giza, Egypt
R.M. Shaapan
Department of Zoonosis, Veterinary Research Division, National Research Center, Dokki, Giza, Egypt