Research Article
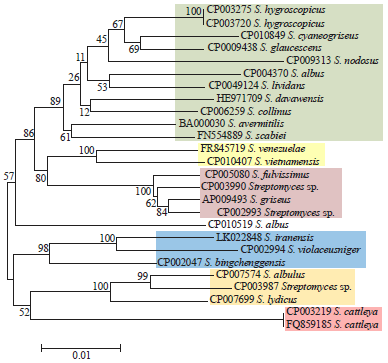
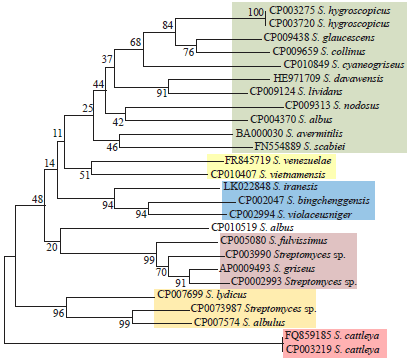
Identification and Phylogeny of Streptomyces Based on Gene Sequences
Universidade do Estado do Amazonas (UEA), Escola Superior de Ciencias da Saude, Pos-Graduacao em Biotecnologia E Recursos Naturais, Manaus, Amazonas, Brasil
Cinthya Paola Ortiz Ojeda
Universidade do Estado do Amazonas (UEA), Escola Superior de Ciencias da Saude, Pos-Graduacao em Biotecnologia E Recursos Naturais, Manaus, Amazonas, Brasil
Ingrid Reis da Silva
Universidade Federal do Amazonas (UFAM)-Programa Multi-Institucional de Pos-Graduacao em Biotecnologia, Manaus, Amazonas, Brasil
Rudi Emerson de Lima Procopio
Universidade do Estado do Amazonas (UEA), Escola Superior de Ciencias da Saude, Pos-Graduacao em Biotecnologia E Recursos Naturais, Manaus, Amazonas, Brasil
LiveDNA: 55.16290