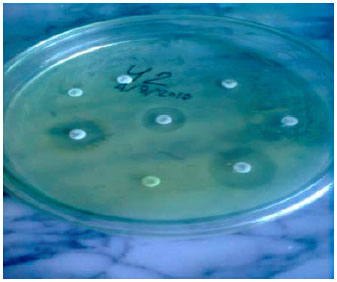
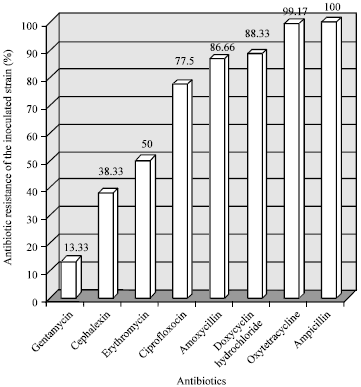
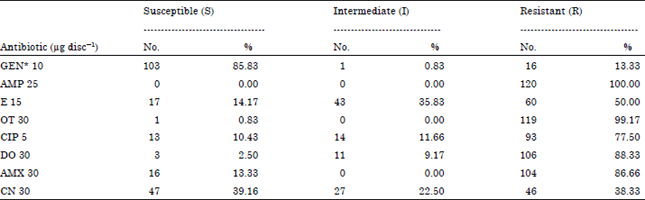
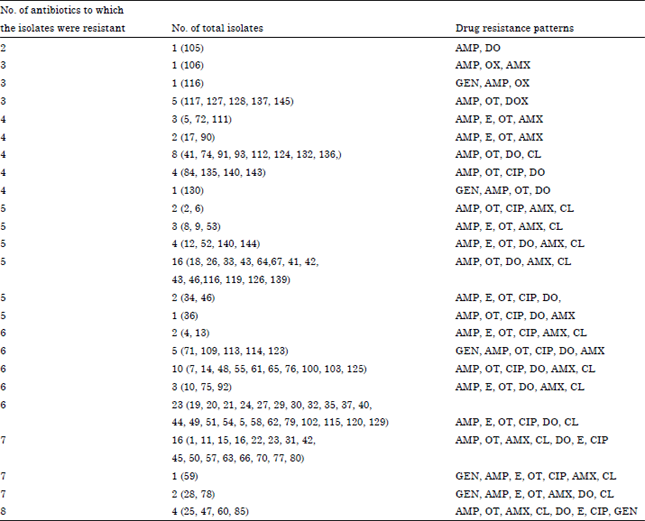
Research Article
Detection of Staphylococcus aureus in Frozen Chicken Rinse through Bacteriological and Nuc Gene Specific PCR Methods and their Drug Resistance Patterns in Southern Chittagong, Bangladesh
Department of Genetic Engineering and Biotechnology, University of Chittagong, Chittagong, 4331, Bangladesh
Mahmuda Akter
Department of Genetic Engineering and Biotechnology, University of Chittagong, Chittagong, 4331, Bangladesh
Zinat Farzana
Department of Genetic Engineering and Biotechnology, University of Chittagong, Chittagong, 4331, Bangladesh
Abdul Jabber Bin Kader
Department of Genetic Engineering and Biotechnology, University of Chittagong, Chittagong, 4331, Bangladesh
Inkeyas Uddin
Department of Genetic Engineering and Biotechnology, University of Chittagong, Chittagong, 4331, Bangladesh
A.M.A.M. Zonaed Siddiki
Department of Genetic Engineering and Biotechnology, University of Chittagong, Chittagong, 4331, Bangladesh
K.M. Kamaruddin
Department of Genetic Engineering and Biotechnology, University of Chittagong, Chittagong, 4331, Bangladesh