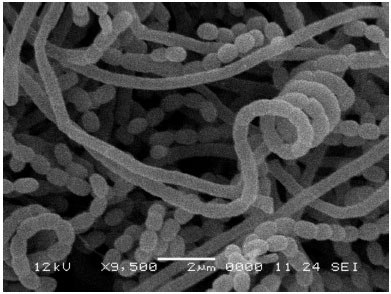
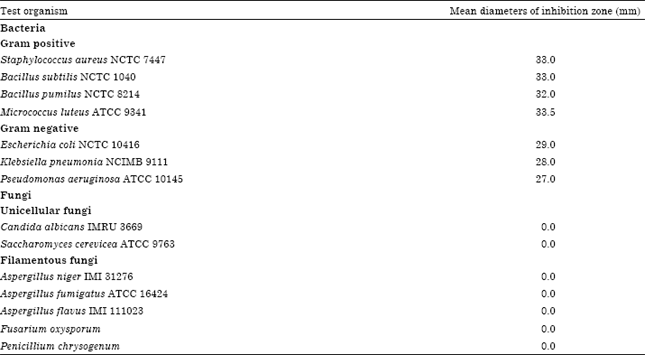
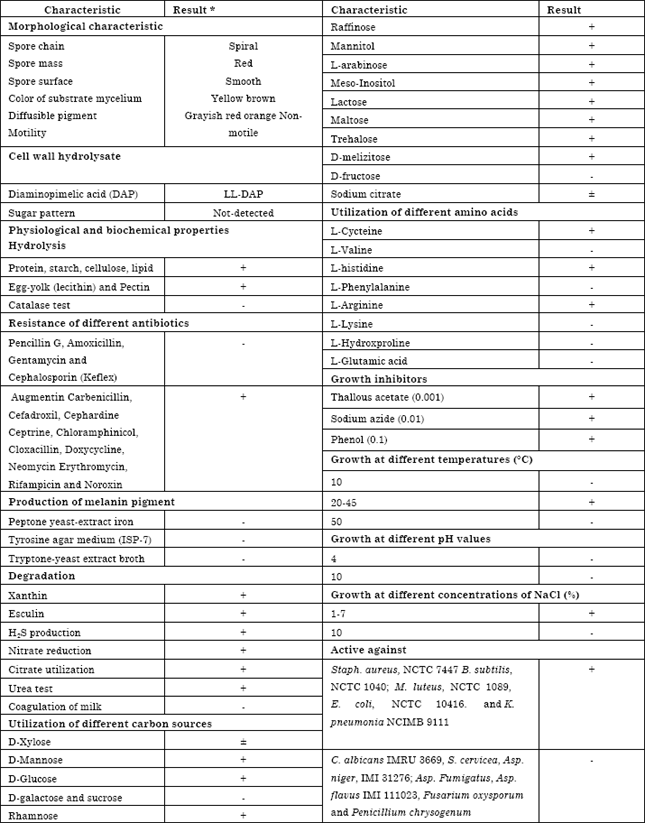
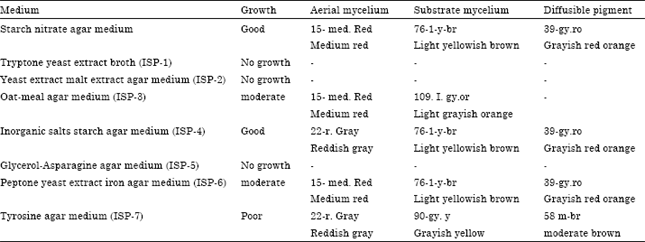
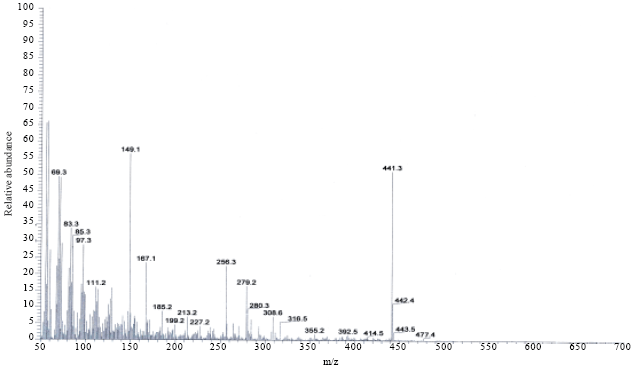
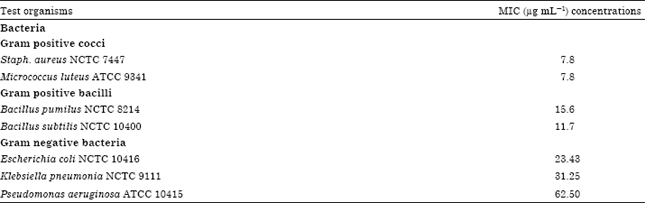
Research Article
Microbiological Studies on the Production of Antimicrobial Agent by Actinomycete Isolated from Saudi Arabia
Department of Botany and Microbiology, Faculty of Science, Al-Azhar University, Assuit, 71524, Egypt
Houssam M. Atta
Department of Botany and Microbiology, Faculty of Science (Boys), Al-Azhar University, Cairo, Egypt
Reda A. Bayoumi
Department of Biotechnology, Faculty of Science and Education, Al-Khurmah Branch, Taif University, KSA