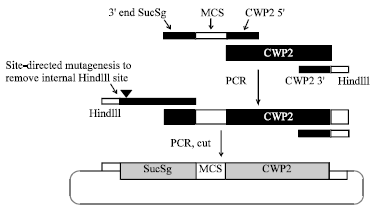
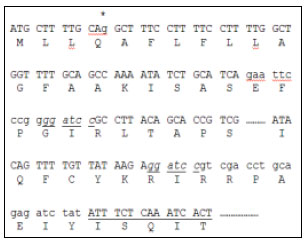
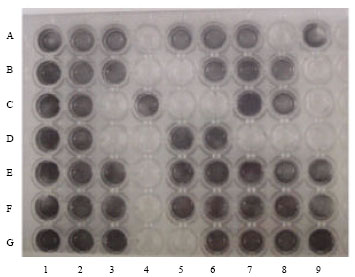
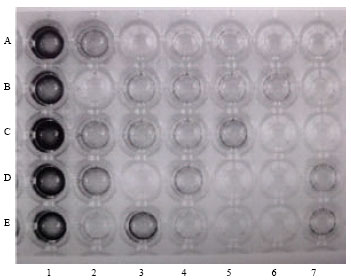
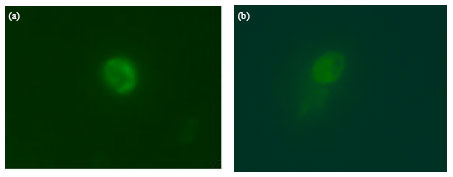
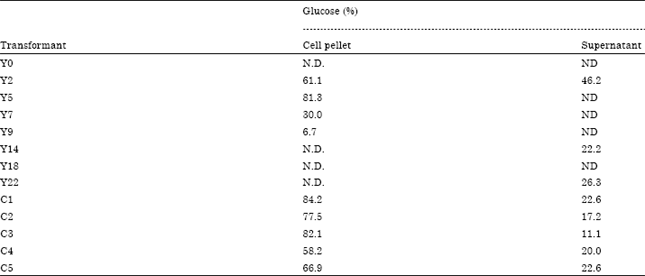
Research Article
Construction of a Vector for the Surface Display of Heterologous Proteins in Saccharomyces cerevisiae
Faculty of Applied Sciences, Universiti Teknologi MARA, 40450 Shah Alam, Selangor, Malaysia
Z.M. Noor
Faculty of Applied Sciences, Universiti Teknologi MARA, 40450 Shah Alam, Selangor, Malaysia
M.F.F. Abdullah
Faculty of Applied Sciences, Universiti Teknologi MARA, 40450 Shah Alam, Selangor, Malaysia