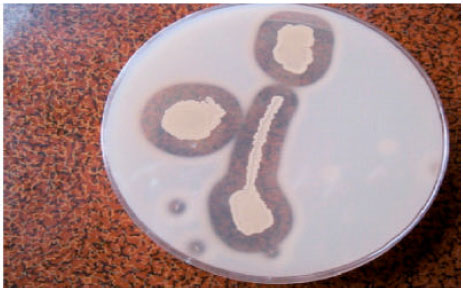
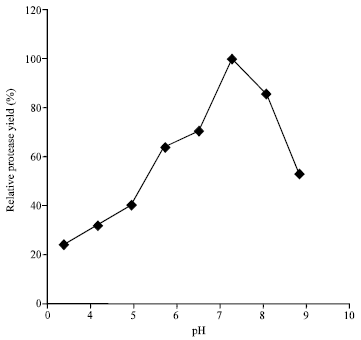
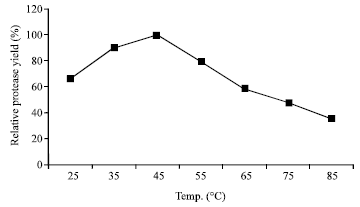
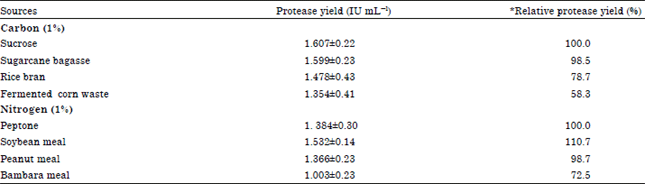
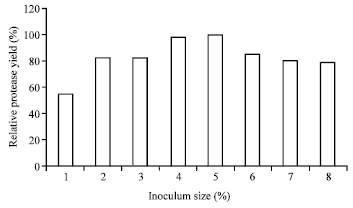
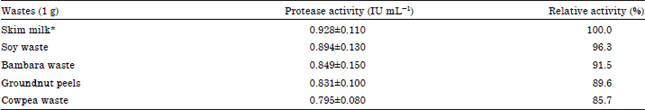
Research Article
Screening and Optimal Protease Production by Bacillus sp. Sw-2 Using Low Cost Substrate Medium
Department of Applied Microbiology and Brewing, Enugu State University of Science and Technology, Enugu, Nigeria
E.E. Mike-Anosike
Department of Microbiology, Tansian University, Umunya, Anambra State, Nigeria