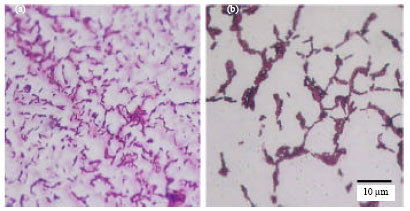
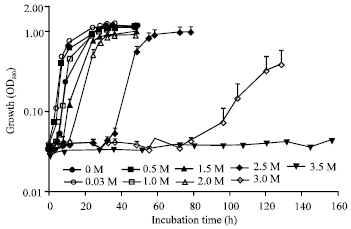
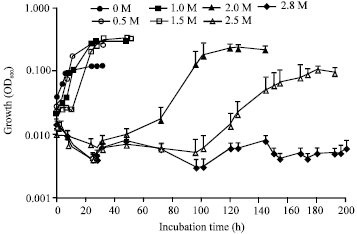
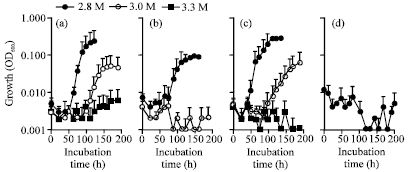
Research Article
Isolation of Extreme Halotolerant Bacteria from Asian Desert Dust; Molecular Phylogeny and Growth Properties of their Cells
College of Science and Engineering, Iwaki Meisei University, Iwaki, Fukushima 970-8551, Japan
E. Iwata
College of Science and Engineering, Iwaki Meisei University, Iwaki, Fukushima 970-8551, Japan
A. Oshima
Faculty of Life and Environmental Science, Shimane University, Matsue, Shimane 690-8504, Japan
A. Ishida
Graduate School of Science and Technology, Kumamoto University, Kumamoto 860-8555, Japan
S. Nagata