Research Article
A Real-Time Polymerase Chain Reaction Based Assay for the Detection of Escherichia coli in Patients with Urinary Tract Infection in the Sudan
College of Medical Laboratory Science, Sudan University of Science and Technology, Sudan
Zahra K. Yousif
National Health Laboratory, Khartoum, Sudan
Mugahid M. El Hassan
College of Medical Laboratory Science, Sudan University of Science and Technology, Sudan
Misk El Yamen A. Atti
College of Medical Laboratory Science, Sudan University of Science and Technology, Sudan
Mansour M. Mansour
College of Medical Laboratory Science, Sudan University of Science and Technology, Sudan










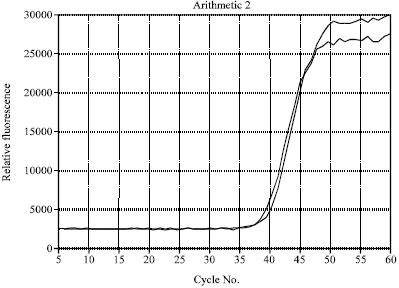
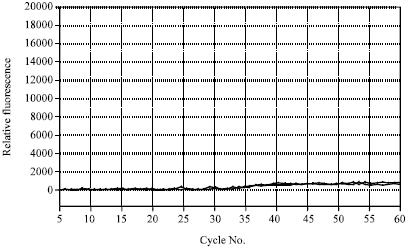
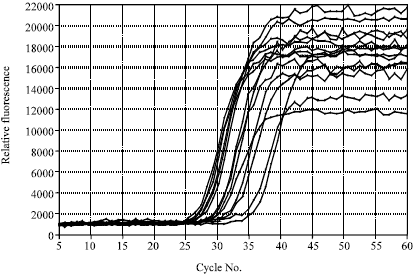


Zarizal Suhaili Reply
This article/ journal can be more specific if the author proceed the detection scheme by differentiating each spp. as well as genus with melting curve analysis instead of using amplification plots alone.
thank you.