Research Article
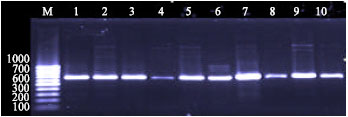
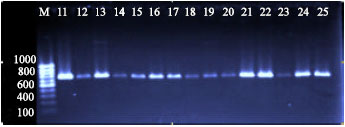
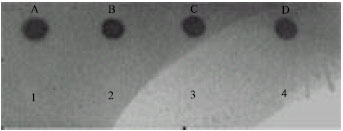
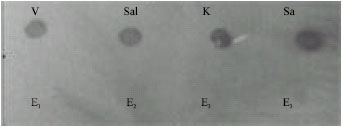
Development of Polymerase Chain Reaction and Dot Blot Hybridization to Detect Escherichia coli Isolates from Various Sources
Department of Microbiology and Parasitology, Faculty of Medicine and Heath Sciences,
N. S. Mariana
Department of Microbiology and Parasitology, Faculty of Medicine and Heath Sciences,
A. R. Raha
Department of Bioprocess Technology, Faculty of Biotechnology and Biomolecular Sciences, University Putra Malaysia, 43400 Serdang, Selangor, Malaysia
Z. Ishak
Biotechnology Center, Mardi, P.O. Box 12301, Malaysia