Research Article
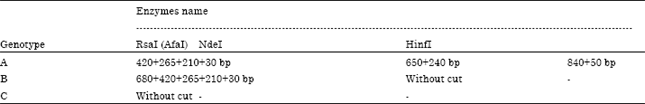
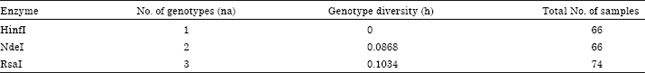
Polymorphism Analysis of Mitochondrial DNA Control Region of Hawksbill Turtles (Eretmochelys imbricata) in the Persian Gulf
Department of Genetics, Faculty of Science, Shahid Chamran University of Ahvaz, Ahvaz, Islamic Republic of Iran
E. Modheji
Department of Genetics, Faculty of Science, Shahid Chamran University of Ahvaz, Ahvaz, Islamic Republic of Iran
H. Zolgharnein
Department of Marine Biology, Faculty of Marine Science, Khorramshahr University of Marine Science and Technology, Khorramshahr, Islamic Republic of Iran