ABSTRACT
Population study focusing the genetic diversity had been widely exploited for wild species but unfortunately little attempt was being made to understand the dynamics of genetic diversity on cultured aquarium species. This study was carried out to understand the dynamics of genetic diversity of cultured live-bearer fish in Malaysia. Taxonomic groupings of both Xiphophorus and Poecilia spp was validated using molecular markers. One hundred and thirty one samples of locally cultured live-bearer fishes which comprised of Xiphophorus maculatus, Xiphophorus helleri, Poecilia reticulata and Poecilia latipinna were sampled. The live-bearer fish samples were genotyped with 24 microsatellite (SSRs) primer pairs. Fifteen out of 24 SSRs primer pairs were able to amplify among the live-bearer fishes and were polymorphic. The average number of alleles was low (1 to 8). Differences between mean observed (HO) and expected (He) heterozygosity for the four species live-bearer fishes were small ranging from 0.02 to 0.06 suggesting no drastic reduction of heterozygosity in cultured fish. The pair-wise comparisons of FST between these four species of live-bearer in general was high (FST>0.25). Such observation was expected as the four tested species of live-bearer belong to different taxonomic groups. A clear distinction between the species of Xiphophorus and Poecilia as revealed by SAHN-clustering tree plot seems to be in accordance with their taxonomy. The data generated from this study provide useful information in understanding the levels of genetic variation in cultured aquarium fish for resource management and conservation.
PDF Abstract XML References Citation
How to cite this article
DOI: 10.3923/jfas.2012.183.193
URL: https://scialert.net/abstract/?doi=jfas.2012.183.193
INTRODUCTION
Molly belongs to the group of live-bearer fish from the genus Poecilia and subgenus Mollienesia (Miller, 1975) which was originated from North America and Central America and distributed to various countries including Malaysia. It was first introduced into Malaysia fifteen years ago and had been locally cultured at Ulu Tiram, Johore for trading. According to a recent report by Department of Fisheries Malaysia (DOF, 2010), this species of live-bearer is one of the major export fish species in the country which consists of Poecilia latipinna, Poecilia velifera and Poecilia sphenops. However, there is no report on the presence of wild species in nature to date in the country.
A wide range of studies had been carried out to understand the genetic diversity dynamics on wild aquatic species such as in planktonic shrimps, prawns, pikeperch and sturgeon (Aziz et al., 2010; Gharibkhani et al., 2009; Norouzi et al., 2008). However, limited studies were attempted on cultured aquarium species. Intensive captive culture practices for a long period of time might have an effect on the overall genetic diversity in any fish species. Thus, it will be beneficial to understand the dynamics of genetic diversity in cultured aquarium species as such information could greatly facilitate choices of selection in any breeding program or in other downstream activities/studies particularly in development of mapping population.
Other fish species that belong to the group of live-bearer include platy (Xiphophorus maculatus), swordtail (Xiphophorus helleri) and guppy (Poecilia reticulata). Taxonomy had assigned molly in the same genus as guppy (both belongs to the genus of Poecilia). Studies by Meyer et al. (1994) had validated taxonomy grouping of both the Xiphophorus species with P. reticulata. Specifically, phylogenetic tree revealed that both X. maculatus and X. helleri were closely related and shared the same cluster while P. reticulata appeared to be an outgroup taxon. On the other hand, a separate study had been carried out by Ptacek and Breden (1998) on several Poecilia species within the subgenus Mollienesia. The sailfin molly (P. latipinna) was found to be closely related to the shortfin molly groups (P. sphenops and P. mexicana). They also observed that guppy (P. reticulata) showed high divergence and was not included within the Mollienesia taxon. Molecular validation on the taxonomic relationships between sailfin molly (P. latipinna) and the other three species of the live-bearer had not been studied so far.
Cross amplification of microsatellite (SSRs) markers between closely rela ted species is common and had been widely used in phylogeny and population genetics (Chistiakov et al., 2006). Although, the use of microsatellite markers in phylogenic studies is not as informative as other markers however, data generated from these markers are increasing (Goldstein et al., 1999; Heath et al., 2001; Reusch et al., 2001). In the study, of population dynamics, SSRs had been widely applied due to their codominant character, small length, extensive genome coverage, relative abundance and high mutation rate. Since SSRs evolved fast, they are powerful tool for analyzing recent and contemporary event (Ellegren, 2000).
Hence, this study was carried out to understand the dynamics of genetic diversity in cultured aquarium species and to validate taxonomic groupings of both the Xiphophorus and Poecilia species by applying SSRs markers.
MATERIALS AND METHODS
Fish collection: Fish were collected from Aquatic International, Subang Jaya, Malaysia in the middle of October 2009 and maintained at Aquaculture Research Centre, Universiti Putra Malaysia, Puchong. These fishes originated from USA and had been locally cultured at Ulu Tiram, Johore, Malaysia. A total of 31 samples of red platy (X. maculatus), 32 samples of green swordtail (X. helleri), 30 samples of cultured assorted guppy (P. reticulata) and 38 samples of white sailfin molly (P. latipinna) were randomly selected. All the samples used were adults of approximately more than three months old with size range between 4.5 to 5.5 cm in both the platy and swordtail, 3.0 to 4.0 cm in guppy and 4.5 to 6.5 cm in molly. Each species of the live-bearer fish was designated as ‘species group’ throughout the text.
DNA extraction and microsatellite genotyping: Genomic DNA was extracted from fresh caudal fin clips according to kit protocols from Genomic Wizard Extraction Kit (Promega) with minor modification.
| Table 1: | Characteristics of fifteen microsatellite prime pairs developed for Xiphophorus maculatus (Walter et al., 2004) tested on the four live-bearer species |
 | |
A total of 26 microsatellite (SSRs) primer pairs designed by Walter et al. (2004) for X. maculatus were downloaded from online database (http://www.xiphophorus.txstate.edu/research/xiphbase/microsat.html) (Table 1). These primer pairs had been tested and successfully amplified on Poecilia species (Walter et al., 2004). A Polymerase Chain Reaction (PCR) amplification was carried out using Eppendorf (Mastercycler gradient) and the PCR reaction volumes was reduced to 10 μL and thermal cycling conditions were carried according to the description by Promega product information (Catalog No. M8295). The amplified PCR products were electrophoresed on 4% metaphor gel (1X TBE buffer, 0.5 μg mL-1 gel red). Scoring of alleles was estimated using Promega ladder catalog number (No. G4511).
Data analysis: Microsatellite genotypes were submitted to MICROCHECKER (Van Oosterhout et al., 2004) for null allele detection. Species group with null alleles were adjusted accordingly. Adjusted genotype frequency were analyzed with GENEPOP version-4 (Raymond and Rousset, 1995) for estimation of genetic diversity which comprised of the number of alleles and the observed and expected heterozygosity (HO and HE) at each locus. Polymorphic Information Content (PIC) of each microsatellite loci was calculated based on formula by Botstein et al. (1980) using Excel Microsatellite Tool kit (Park, 2001). Conformation to Hardy Weinberg Equilibrium (HWE) was carried out by locus and by population using POPGENE32 (Yeh and Boyle, 1997). The pair-wise genetic differentiation which determines the presence of species group structuring was evaluated through F-statistics (FIS, FIT and FST) (Weir and Cockerham, 1984). F-STAT (Goudet, 1995) was used to estimate FST (Weir and Cockerham, 1984) and RST (Slatkin, 1995) across species groups. The genetic disequilibrium between each pair of loci was estimated using GENEPOP version 4 (Raymond and Rousset, 1995). The genetic distance which measures the amount of variation between pairs of species group was calculated through isolation by distance between groups fitting in the equation of Dσ2 = FST (1-FST) (Rousset, 1997). The NTSYS version-2.1 (Rohlf, 1998) was subsequently used to construct a tree plot employing SAHN-clustering calculation.
RESULTS
Genetic diversity and population structuring: Fifteen out of 26 SSRs primer pairs were able to amplify within and among species groups. Null alleles were present in all the loci even after genotypes adjustment with the tendency towards homozygote excess. However, there was no scoring error due to stuttering and no evidence of large allele dropout in all the loci evaluated. Three out of 15 SSRs primer pairs namely Msa018, Msa035 and Msa032 could not amplified on P. reticulata species. A total number of 170 alleles were detected. The highest total number of allele was found in P. latipinna (58 alleles) and the lowest was found in X. maculatus (30 alleles) (Table 2).
The P. latipinna showed highest number of primer pairs which were informative having Polymorphic Information Content (PIC) values more than 0.5 as compared with other species groups. Seven out of 15 primer pairs fell in informative category namely Msa120 (PIC = 0.74), Msb036 (0.51), Msa090 (0.62), Msa061 (0.67), Msa077 (0.77), Msa108 (0.59) and Msb023 (0.58). Majority of PIC values in X. maculatus fell in less informative category (PIC<0.5) with an exception in primer pair Msb023. In X. helleri, only primer pairs Msa069 and Msa032 were categorized as informative with the PIC values of 0.53 and 0.57, respectively. Similarly, two out of the 15 primer pairs in P. reticulata fell in the informative categories which were Msb023 and Msa026 having PIC values of 0.60 and 0.51, respectively (Table 2).
Departures from HWE was significant (p<0.05) in most of the primer pairs even after genotype adjustment. In X. maculatus, only three primer pairs confirmed to HWE. In X. helleri, six primer pairs were in HWE. The P. reticulata recorded the highest number of primer pairs that confirmed to HWE with a total of eight primer pairs while in P. latipinna, only four primer pairs were in HWE. On average, the HWE results were not in equilibrium in the four species groups evaluated (Table 2).
The average observed heterozygosity (HO) ranged from 0.05 to 0.27 while, the average expected heterozygosity (He) ranged from 0.07 to 0.27. The P. latipinna had the highest average HO among the four species groups while the lowest was found in X. maculatus. Overall, both X. helleri and P. reticulata showed mean HO higher than HE. The FIS values on both species were negative, indicating heterozygote excess. On the other hand, X. maculatus showed lower mean HO compared to the HE. A check on the FIS value was positive, indicating heterozygote deficiency. The values of HO and HE were similar in P. latipinna (Table 3).
In population genetics, a test for Linkage Disequilibrium (LD) investigates genotypic disequilibrium at different set of primer pairs. In other words, LD is to test association of alleles by different sets of primer pairs. Overall, LD was observed between thirty-five out of 105 possible combinations (approximately 33%). However, only primer pair Msc029 did not show linkage disequilibrium with all the other primer pairs with an exception with primer pair Msb023 (Table 4).
The average estimator of genetic differentiation using RST was higher than of estimated from FST across the species group (Table 5). Pair wise comparisons in FST were high (FST>0.25) ranging from 0.33 to 0.86. The highest pairwise FST value was found between X. maculatus and P. latipinna and the lowest was found between P. reticulata and X. helleri (Table 6). The largest difference in genetic distance was found between X. maculatus and P. latipinna and the smallest was found between P. reticulata and P. latipinna (Table 6). The SAHN-clustering tree plot generated by utilizing genetic distance values revealed two clusters. The first cluster comprised of X. maculatus and X. helleri while the second cluster was shared by both P. reticulata and P. latipinna (Fig. 1). These four species of live-bearer groups seem to be clustered according to their respective genus that corroborates their taxonomy.
| Table 2: | Microsatellite markers, number of alleles (Na), polymorphic information content (PIC) and probability in Hardy-Weinberg equilibrium (HWE) |
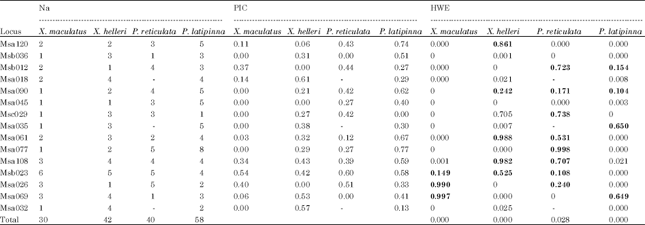 | |
| Values in bold showed SSRs primer pairs in HWE balance; 0 monomorphic:- No amplification | |
| Table 3: | Microsatellite markers, observed (Ho) and expected (HE) heterozygosity and F-statistics (FS values were computed as described by Weir and Cockerham (1984) (W and C) |
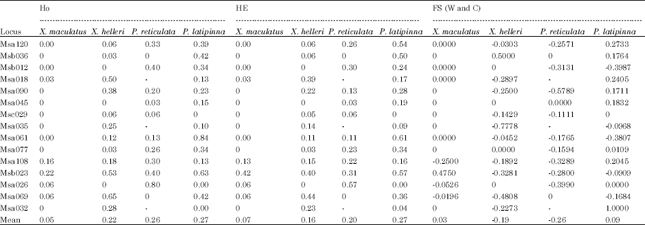 | |
| 0: Monomorphic; -: No amplification | |
| Table 4: | Pairwise Linkage Disequilibrium (LD) between 15 microsatellite primer pairs |
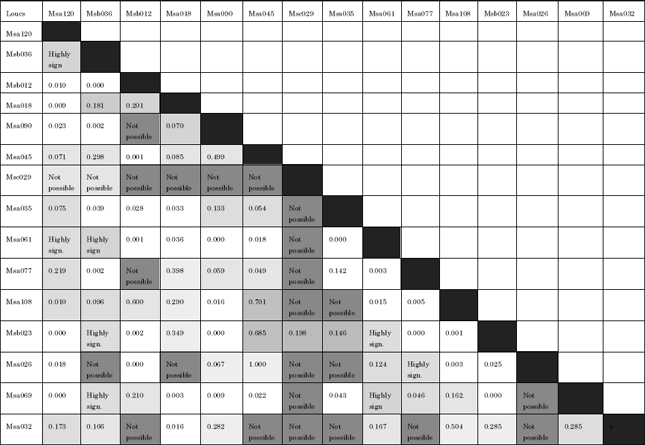 | |
| Light gray: Significant association between markers (p<0.05). Black highlighted: No possible association. White boxes: Not significant | |
| Table 5: | Overall and per locus FST and RST values for four different species of live-bearer fish |
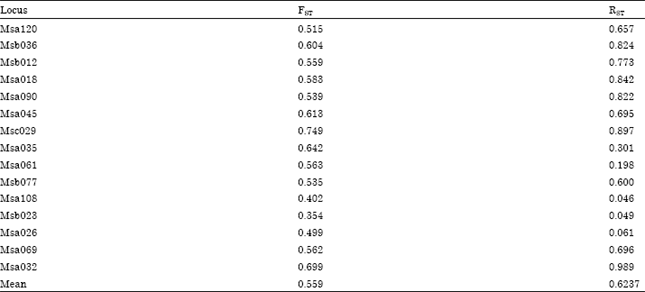 | |
| Table 6: | Pair-wise comparisons among species |
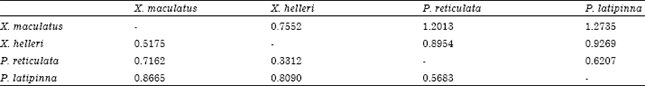 | |
| Values below diagonal are estimates of pairwise FST and values above diagonal are estimates of genetic distance fitting in the equation of Dσ2 = FST (1-FST) (Rousset, 1997) | |
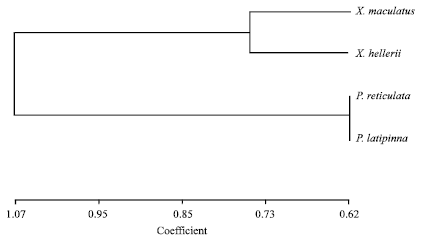 | |
| Fig. 1: | SAHN-clustering tree plot based on pairwise genetic distance (Dσ2) on four species of live- bearer fishes available in Malaysian market |
DISCUSSION
The allelic diversities were low (mean number of alleles ranged from 1 to 8) in the four live bearer fish evaluated. Such observation is expected in cultured fish. Similar observation was found by Norris et al. (1999) in farmed Atlantic salmon which showed lower diversity in comparison with those from the wild. Possible explanation for such occurrence would be small population size, genetic drift, geographical isolation and limited gene flow (Lowe et al., 2004; Norris et al., 1999). Genetic drift due to geographical isolation or migration will limit the gene flow and subsequently causing low allelic diversity in a newly colonized population. However, such events are more likely to occur in wild population. Another possible reason is artificial selection. As, most of the sample taken in this study was largely cultured, human intervention in selecting fish having favorable traits in breeding will eventually leads to reduction in allele diversity.
Although, there were differences between HO and HE in all the four species of live-bearer with an exception on P. latipinna, the value of differences were small ranging from 0.02 to 0.06. On average, both X. helleri and P. reticulata showed HO higher than HE, theoretically suggesting heterozygous excess. Possible explanation on the excess of heterozygosity in this study might be due to the type of breeding practices carried out in cultured aquarium fish. Hybridization by crossbreeding to enhance colour and appearance might be causal factor that increased the heterozygosity level on both of the species groups. There is no sign of inbreeding or outbreeding in P. latipinna detected as both HO and He values were similar Fifteen years of captive selective breeding either through inbreeding or crossbreeding might not be long enough to contribute significantly to any changes in heterozygosity level (Anderson and Hayes, 2005). This study did not detect any drastic heterozygosity reduction in these four species of live-bearer despite being a cultured fish species. This result was in agreement with the findings by Norris et al. (1999) which similarly did not show reduction of heterozygosity in farmed fish. This study also demonstrates that low allelic diversity does not have a direct correlation with the level of heterozygosity. Similar observation was also found by Norris et al. (1999) which stated that decrease in allelic diversity is not detectable from levels of heterozygosity.
The average estimator of genetic differentiation across the species group shown by RST was higher than FST. Such observation was in agreement with Slatkin (1995), which stated that FST tends to understimatesthe true level of genetic differentiation. The pair-wise comparison of FST was high (FST>0.25) between the four species of live-bearers. Such finding was expected as the four species of live-bearer under study belongs to different taxonomic groups. The highest level of genetic differentiation was found between P. latipinna and X. maculatus followed by X. helleri. Lower level of genetic differentiation was found between P. latipinna and P. reticulata. This result was expected as closely related taxa generally exhibit lower level of genetic differentiation between them than distantly related taxa (Lowe et al., 2004).
The SAHN clustering tree plot showed that both X. maculatus and X. helleri were grouped in the same cluster while both P. reticulata and P. latipinna were grouped in another cluster. The clustering pattern of both Xiphophorus and Poecilia species as revealed by molecular markers seems to be in accordance with their taxonomy. This finding is in agreement with phylogenetic studies by Meyer et al. (1994) which showed that X. helleri appeared to be the sister group of X. maculatus and P. reticulata appeared to be an outgroup taxon. However, P. latipinna species group was not included in the study by Meyer et al. (1994) and made such comparison impossible.
CONCLUSION
The microsattellites (SSRs) markers applied in this study had successfully revealed low allelic diversity in these four cultured species of live bearer fishes. The grouping of both Xiphophorus and Poecilia species revealed by molecular marker in this study seem to be in accordance with their respective genus that corroborates with their taxonomy. Thus the data gathered from this study provide baseline information in understanding genetic variation of cultured aquarium fish for resource management and conservation.
ACKNOWLEDGMENT
Thanks are extended to Department of Aquaculture, Universiti Putra Malaysia and FRGS 01-04-10-825FR grant from Ministry of Higher Education (MOHE), Malaysia for providing the facilities and financial support, respectively.
REFERENCES
- Aziz, D., S.S. Siraj, A. Arshad, S.M.N. Amin and S.A. Harmin, 2010. Population characterization of planktonic shrimp, Acetes japonicus (Decapoda: Sergestidae) using RAPD technique. J. Biol. Sci., 10: 355-361.
CrossRefDirect Link - Botstein, D., R.L. White, M. Skolnick and R.W. Davis, 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet., 32: 314-331.
PubMed - Chistiakov, D.A., B. Hellemans and F.A.M. Volckaert, 2006. Microsatellites and their genomic distribution, evolution, function and applications: A review with special reference to fish genetics Aquaculture, 255: 1-29.
CrossRefDirect Link - Ellegren, H., 2000. Microsatellite mutations in the germline: Implications for evolutionary inference. Trends Genet., 16: 551-558.
CrossRef - Goudet, J., 1995. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Heredity, 86: 485-486.
Direct Link - Gharibkhani, M., M. Pourkazemi, M. Soltani, S. Rezvani and L. Azizzadeh, 2009. Population genetic structure of pikeperch (Sander lucioperca Linnaeus, 1758) in the Southwest Caspian Sea using microsatellite markers. J. Fish. Aquatic Sci., 4: 161-168.
CrossRef - Goldstein, D.B., G.W. Roemer, D.A. Smith, D.E. Reich, A. Bergman and R.K. Wayne, 1999. The use of microsatellite variation to infer population structure and demographic history in a natural model system. Genetics, 151: 797-801.
PubMedDirect Link - Heath, D.D., S. Pollard and C. Herbinger, 2001. Genetic structure and relationships among steelhead trout (Oncorhynchus mykiss) populations in British Columbia. Heredity, 86: 618-623.
CrossRef - Meyer, A., J.M. Morrissey and M. Schartl, 1994. Recurrent origin of a sexually selected trait in Xiphophorus fishes inferred from a molecular phylogeny. Nature, 368: 539-542.
CrossRef - Miller, R.R., 1975. Five new species of Mexican poeciliid fishes of the genera Poecilia, Gambusia and Poeciliopsis. Occasional Pap. Museum Zool., 672: 1-44.
Direct Link - Norouzi, M., M. Pourkazemi, A. Keyvan, S.M.R. Fatemi and B. Kazemi, 2008. Population genetic structure of stellate sturgeon (Acipenser stellatus Pallas, 1771) in the South Caspian Sea using microsatellite markers. J. Fish. Aquatic Sci., 3: 158-166.
CrossRefDirect Link - Norris, A.T., D.G. Bradley and E.P. Cunningham, 1999. Microsatellite genetic variation between and within farmed and wild Atlantic salmon (Salmo salar) populations. Aquaculture, 180: 247-264.
CrossRef - Van Oosterhout, C., W.F. Hutchinson, D.P.M. Wills and P. Shipley, 2004. Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes, 4: 535-538.
CrossRefDirect Link - Ptacek, M.B. and F. Breden, 1998. Phylogenetic relationships among the mollies (Poecilidae: Poecilia: Mollienesia group) based on mitochondrial DNA sequences. J. Fish Biol., 53: 64-81.
CrossRef - Raymond, M. and F. Rousset, 1995. GENEPOP (Verison 1.2): population genetics software for exact test and ecumenicism. J. Hered., 86: 248-249.
Direct Link - Reusch, T.B.H., K.M. Wegner and M. Kalbe, 2001. Rapid genetic divergence in postglacial populations of threespine stickleback (Gasterosteus aculeatus): The role of habitat type, drainage and geographical proximity. Mol. Ecol., 10: 2435-2445.
CrossRefDirect Link - Rousset, F., 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics, 145: 1219-1228.
PubMedDirect Link - Slatkin, M., 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics, 139: 457-462.
PubMed - Walter, R.B., J.D. Rains, J.E. Russell, T.M. Guerra and C. Daniels et al., 2004. A microsatellite genetic linkage map for Xiphophorus. Genet. Soc. Am., 168: 363-372.
CrossRef - Weir, B.S. and C.C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution, 38: 1358-1370.
CrossRefDirect Link








