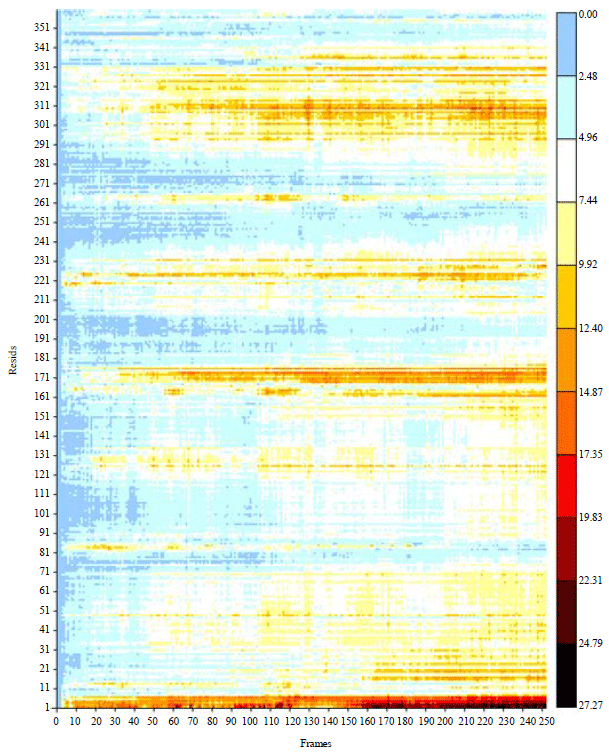
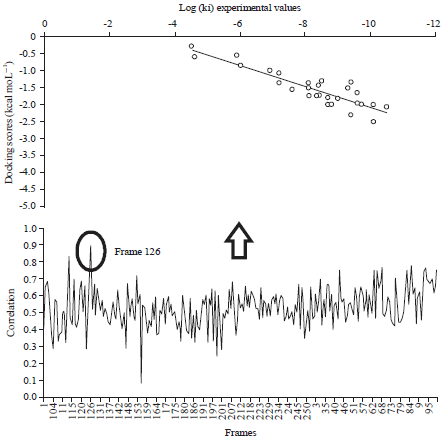
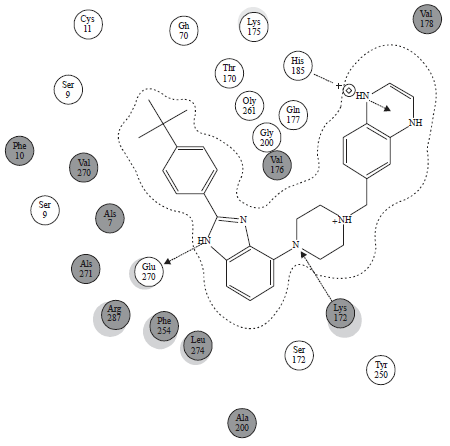
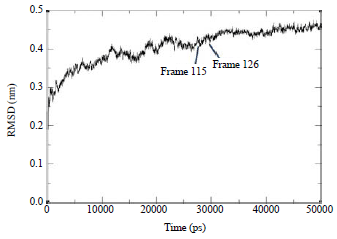
Research Article
Homology Modeling, Molecular Dynamics Simulation and Cross-Docking Studies of Human Histamine-2 (H2) Receptor to Obtain a 3D Structure for Further SBDD Studies
Department of Medicinal Chemistry, Faculty of Pharmacy, Shiraz University of Medical Sciences, Shiraz, Iran
LiveDNA: 98.30889
Hamidreza Ghafouri
Department of Medicinal Chemistry, Faculty of Pharmacy, Shiraz University of Medical Sciences, Shiraz, Iran
Farnaz Salehi
Department of Medicinal Chemistry, Faculty of Pharmacy, Shiraz University of Medical Sciences, Shiraz, Iran
Leila Emami
Department of Medicinal Chemistry, Faculty of Pharmacy, Shiraz University of Medical Sciences, Shiraz, Iran
Neda Khonya
Department of Medicinal Chemistry, Faculty of Pharmacy, Shiraz University of Medical Sciences, Shiraz, Iran
Zahra Rezaei
Department of Medicinal Chemistry, Faculty of Pharmacy, Shiraz University of Medical Sciences, Shiraz, Iran
Amirhossein Sakhteman
Department of Medicine, Faculty of Health Sciences, University of Eastern Finland, Kuopio, Finland
LiveDNA: 98.30944