Research Article
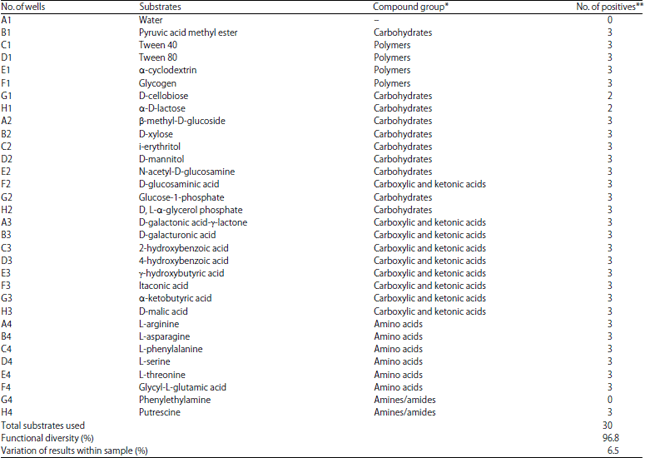
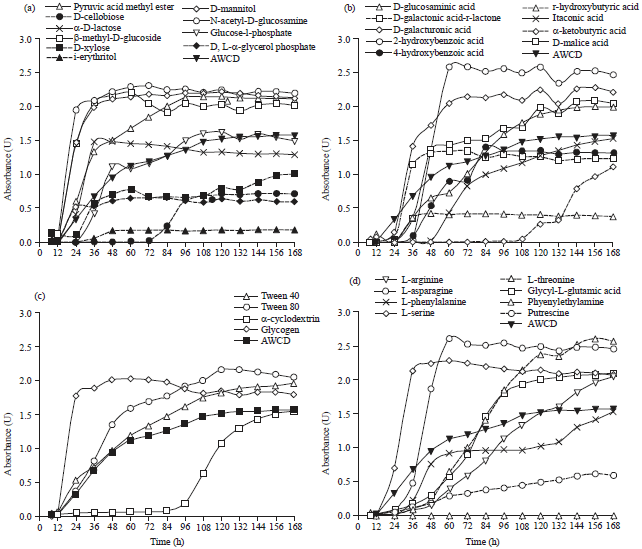
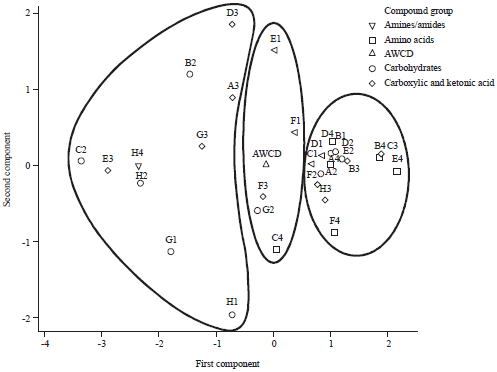
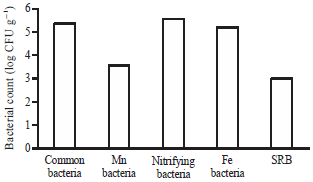
The Ability of Biofilm Community Sampled from Metal Surfaces at Saguling Hydro Power in Utilizing Carbon Sources by Using Biolog EcoPlateTM
Microbial Biotechnology Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, Indonesia
Navisan Najia
Microbial Biotechnology Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, Indonesia
Rahma Widya Ningrum
Microbial Biotechnology Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, Indonesia
Dea Indriani Astuti
Microbial Biotechnology Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, Indonesia
Gede Suantika
Microbial Biotechnology Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, Indonesia
Pingkan Aditiawati
Microbial Biotechnology Research Group, School of Life Sciences and Technology, Institut Teknologi Bandung, Indonesia