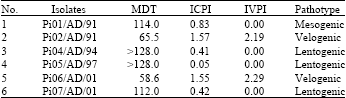
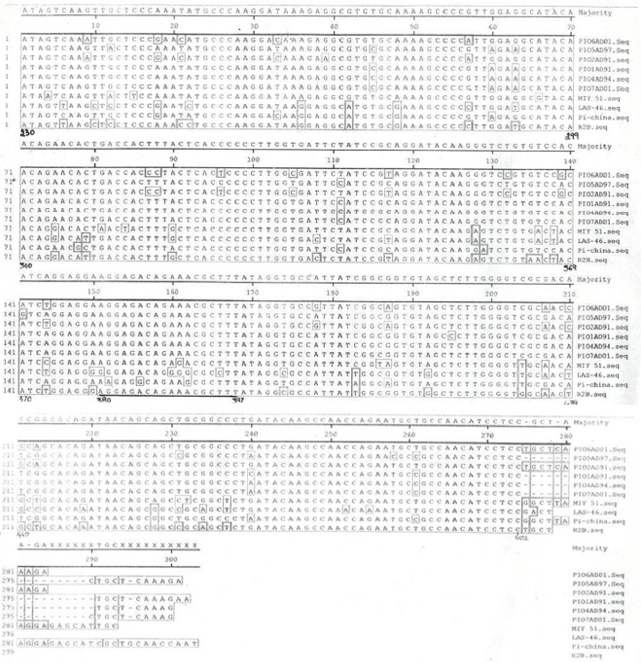
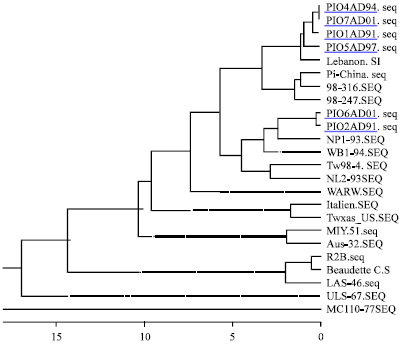
Research Article
Molecular Characterization and Phylogenetic Analysis of Selected Pigeon Paramyxovirus Type-1 (PPMV-1) Indian Isolates
Avian Diseases Section, Division of Pathology, Indian Veterinary Research Institute, Uttar Pradesh, 243 122, India
S.D. Singh
Avian Diseases Section, Division of Pathology, Indian Veterinary Research Institute, Uttar Pradesh, 243 122, India
J.M. Kataria
Central Avian Research Institute, Izatnagar, Uttar Pradesh, 243 122, India
R. Barathidasan
Avian Diseases Section, Division of Pathology, Indian Veterinary Research Institute, Uttar Pradesh, 243 122, India
K. Dhama
Avian Diseases Section, Division of Pathology, Indian Veterinary Research Institute, Uttar Pradesh, 243 122, India