Research Article
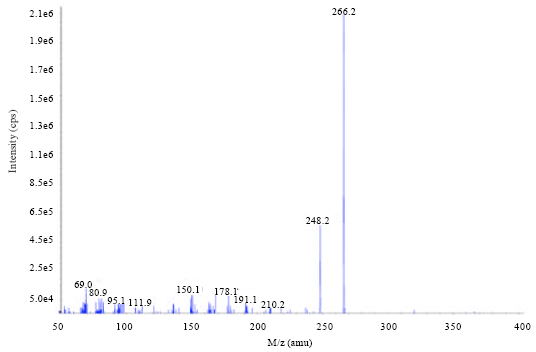
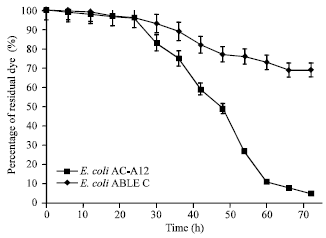
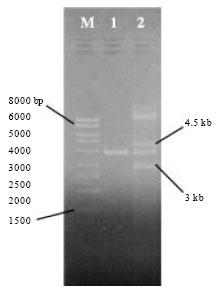
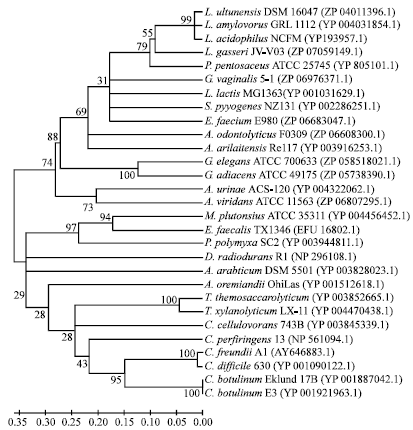
Biosynthesis of Autoinducer-2 as the Possible Mechanism to Enhance Decolourisation of Azo Dye by Citrobacter freundii A1
Nanoporous Materials for Biological Applications Research Group, Sustainability Research Alliance, Universiti Teknologi Malaysia, 81310 UTM Johor Bahru, Johor, Malaysia
Noor Aini Abdul Rashid
Nanoporous Materials for Biological Applications Research Group, Sustainability Research Alliance, Universiti Teknologi Malaysia, 81310 UTM Johor Bahru, Johor, Malaysia
Abdull Rahim Mohd Yusoff
Department of Chemistry, Faculty of Science, Universiti Teknologi Malaysia, 81310 UTM Johor Bahru, Johor, Malaysia
Lee Suan Chua
Institute of Bioproduct Development, Universiti Teknologi Malaysia, 81310 UTM Johor Bahru, Johor, Malaysia