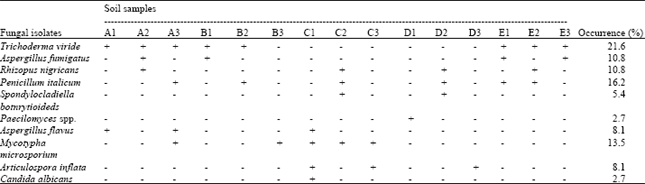
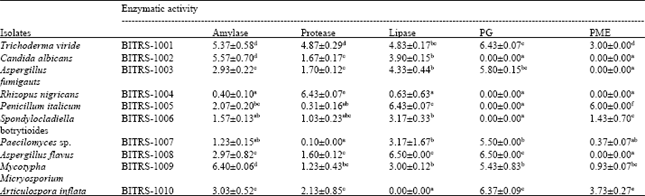
Research Article
Screening of Fungal Isolates from Nigerian Tar Sand Deposit in Ondo State for Novel Biocatalysts
Department of Microbiology, Federal University of Technology, PMB704, Akure, Nigeria
Ogunmolu Funso Emmanuel
Department of Microbiology, Federal University of Technology, PMB704, Akure, Nigeria