Research Article
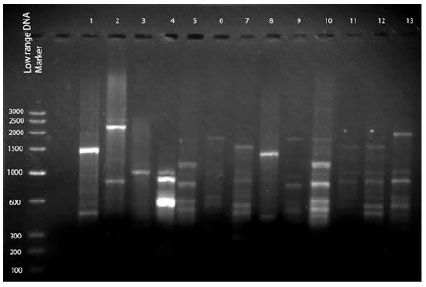
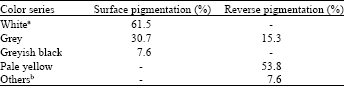
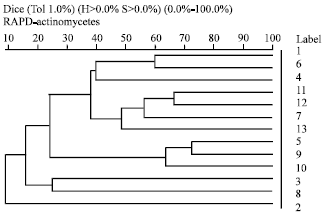
Enzymatic Screening and Random Amplified Polymorphic DNA Fingerprinting of Soil Streptomycetes Isolated from Wayanad District in Kerala, India
Department of Microbiology, Genohelix Biolabs, A Division of Centre for Advanced Studies in Biosciences, Jain University, 127/2, Bull Temple Road, Chamarajpet, Bangalore-560019, India
Arijit Das
Department of Microbiology, Genohelix Biolabs, A Division of Centre for Advanced Studies in Biosciences, Jain University, 127/2, Bull Temple Road, Chamarajpet, Bangalore-560019, India
K. Prashanthi
Department of Molecular Biology, Genohelix Biolabs, A Division of Centre for Advanced Studies in Biosciences, Jain University, 127/2, Bull Temple Road, Chamarajpet, Bangalore-560019, Karnataka, India
Sandeep Suryan
Department of Molecular Biology, Genohelix Biolabs, A Division of Centre for Advanced Studies in Biosciences, Jain University, 127/2, Bull Temple Road, Chamarajpet, Bangalore-560019, Karnataka, India
Sourav Bhattacharya
Department of Microbiology, Genohelix Biolabs, A Division of Centre for Advanced Studies in Biosciences, Jain University, 127/2, Bull Temple Road, Chamarajpet, Bangalore-560019, India