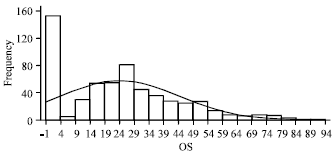
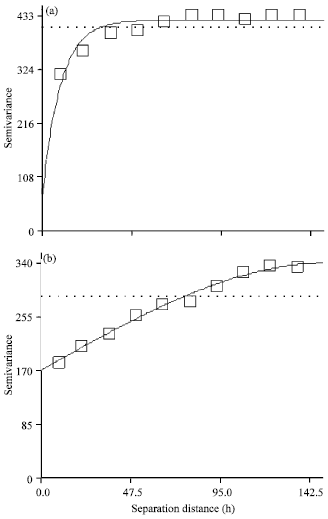
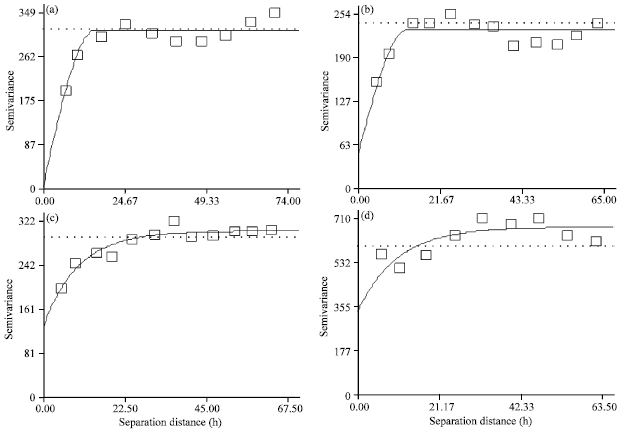
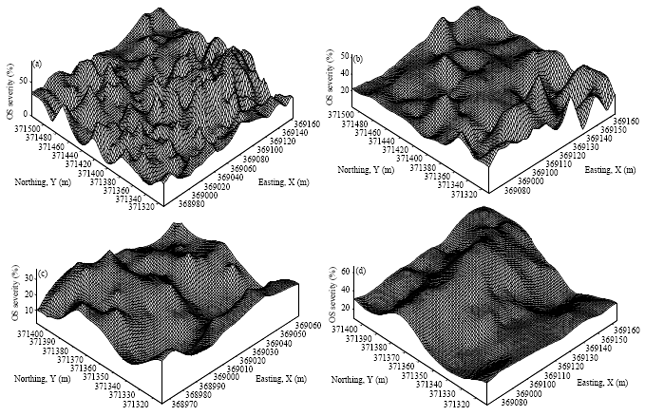
Research Article
Spatial Variability of Orange Spotting Disease in Oil Palm
Department of Agriculture Technology, Universiti Putra Malaysia, 43400 Serdang, Selangor, Malaysia
S.K. Balasundram
Department of Agriculture Technology, Universiti Putra Malaysia, 43400 Serdang, Selangor, Malaysia
G. Vadamalai
Department of Plant Protection, Universiti Putra Malaysia, 43400 Serdang, Selangor, Malaysia
M.H.A. Husni
Department of Land Management, Faculty of Agriculture, Universiti Putra Malaysia, 43400 Serdang, Selangor, Malaysia