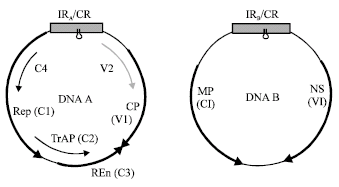
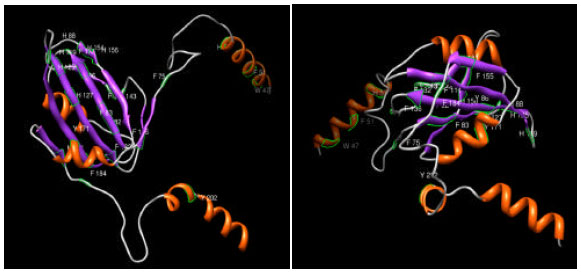
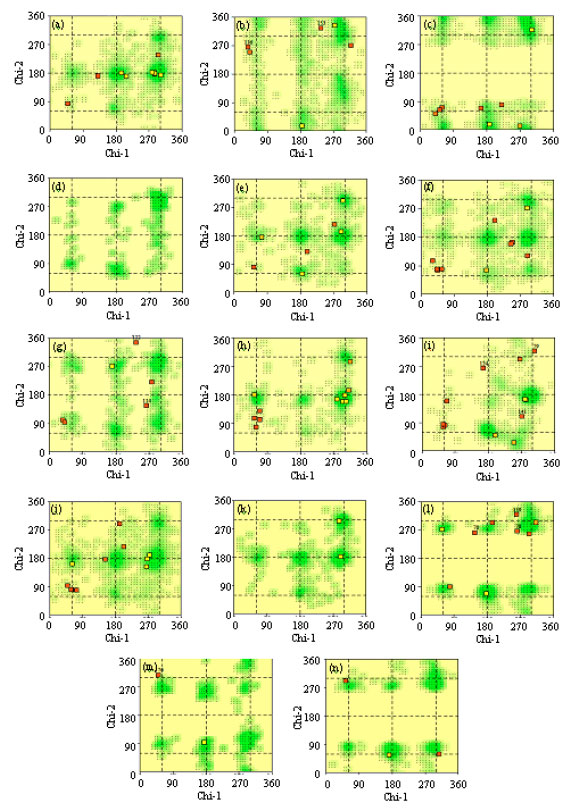
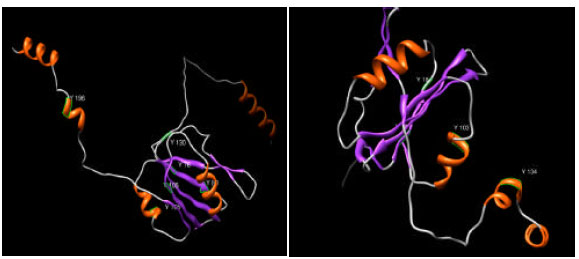
Research Article
In silico Analysis of Genetic Diversity of Begomovirus using Homology Modelling
Department of Biotechnology, MITS University, Lakshmangarh, Rajasthan, India
R.K. Gaur
Department of Biotechnology, MITS University, Lakshmangarh, Rajasthan, India
R. Raizada
Department of Biotechnology, MITS University, Lakshmangarh, Rajasthan, India
V.K. Gupta
Department of Biotechnology, MITS University, Lakshmangarh, Rajasthan, India