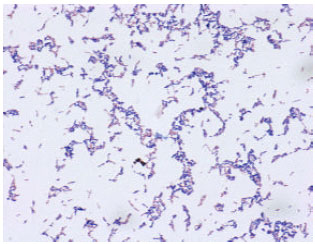
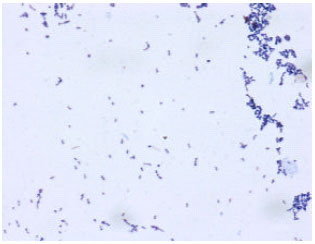
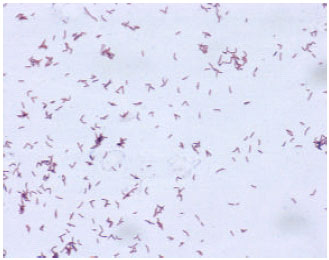
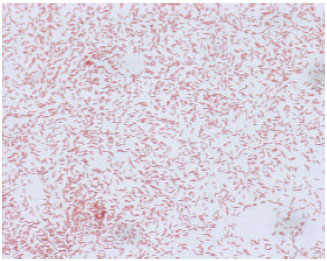
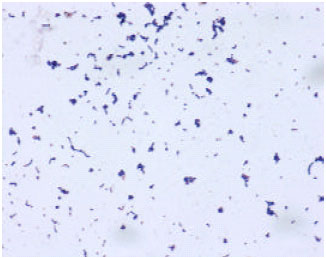
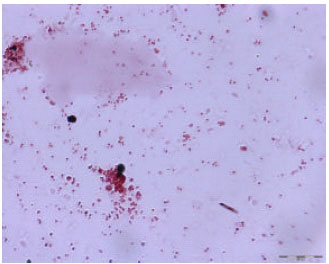
Research Article
Pathogenic Bacteria Predominate in the Oral Cavity of Malaysian Subjects
Division of Microbiology, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia
W.Y. Teoh
Division of Microbiology, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia
S. Muniandy
Department of Molecular Medicine, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia
H. Yaakob
Department of Oral Pathology, Oral Medicine and Periodontology, Faculty of Dentistry, University of Malaya, Kuala Lumpur, Malaysia