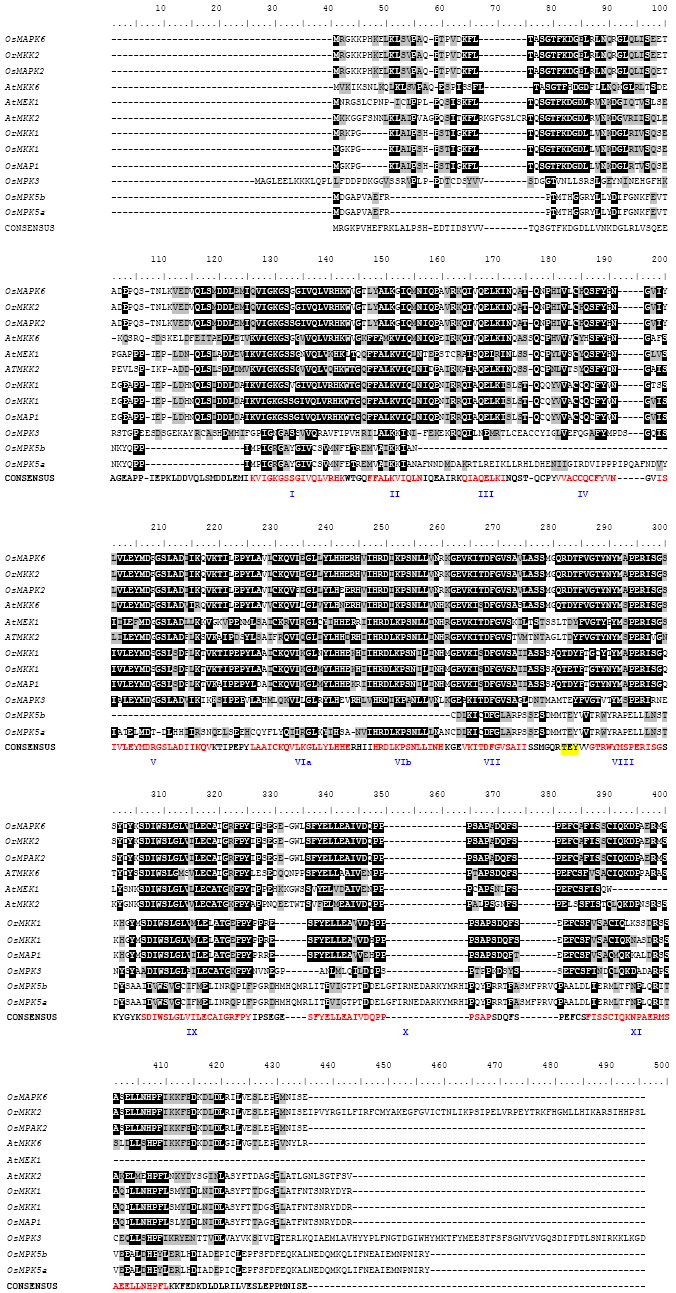
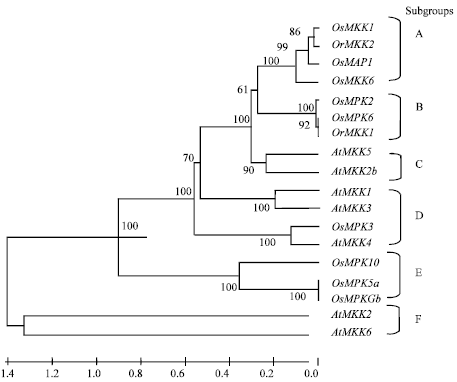
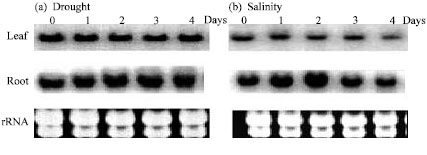
Research Article
The Modulation of Abiotic Stresses by Mitogen-Activated Protein Kinase in Rice
Faculty of Science and Technology, School of Biosciences and Biotechnology, Universiti Kebangsaan Malaysia, 43600 UKM, Bangi, Selangor, Malaysia
N.M. Kasim
Faculty of Science and Technology, School of Biosciences and Biotechnology, Universiti Kebangsaan Malaysia, 43600 UKM, Bangi, Selangor, Malaysia
V.V. Fui
Department of Biotechnology, Faculty of Applied Sciences, UCSI University, No. 1 Jalan Menara Gading. UCSI Heights, 56000 Kuala Lumpur, Malaysia