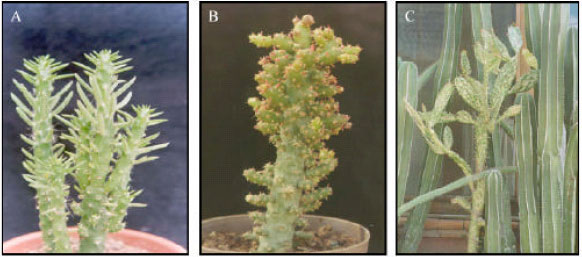
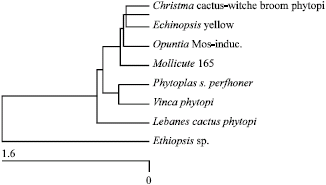
Short Communication
Phytoplasmas Associated to Diseases of Ornamental Cacti in Mexico
Centro de Investigacion y de Estudios Avanzados del Instituto Politecnico Nacional, Campus Guanajuato, km 9.6 Libramiento Norte, Carretera Irapuato-Leon, 36821 Irapuato, Gto., Mexico
F. Parra-Cota
Centro de Investigacion y de Estudios Avanzados del Instituto Politecnico Nacional, Campus Guanajuato, km 9.6 Libramiento Norte, Carretera Irapuato-Leon, 36821 Irapuato, Gto., Mexico
J.C. Ochoa-Sanchez
Centro de Investigacion y de Estudios Avanzados del Instituto Politecnico Nacional, Campus Guanajuato, km 9.6 Libramiento Norte, Carretera Irapuato-Leon, 36821 Irapuato, Gto., Mexico
C. Perales-Segovia
Instituto Tecnologico El Llano Aguascalientes, Apartado Postal 72-4, Administracion Postal 2, Ags., Mexico
J.P. Martinez-Soriano
Centro de Investigacion y de Estudios Avanzados del Instituto Politecnico Nacional, Campus Guanajuato, km 9.6 Libramiento Norte, Carretera Irapuato-Leon, 36821 Irapuato, Gto., Mexico