Research Article
Cloning and Molecular Characterization of Nine α-gliadin Genes from Triticum turgidum ssp. paleocolchicum
Triticeae Research Institute, Sichuan Agricultural University, Yaan, Sichuan 625014, China
Wei Li
Triticeae Research Institute, Sichuan Agricultural University, Yaan, Sichuan 625014, China
Yuming Wei
Triticeae Research Institute, Sichuan Agricultural University, Yaan, Sichuan 625014, China
Guoyue Chen
Triticeae Research Institute, Sichuan Agricultural University, Yaan, Sichuan 625014, China
Youliang Zheng
Triticeae Research Institute, Sichuan Agricultural University, Yaan, Sichuan 625014, China









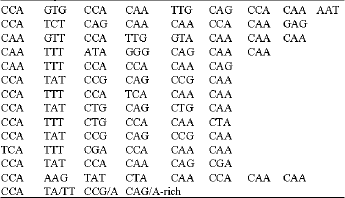 ence of the Gli1-2 repetitive domain is arranged by codons and suggested repeats are arrayed vertically. A consensus structure is given below. The vertical line separates the conserved first three codons of each repeat motif from the variable-length glutamine-rich part of the repeat
ence of the Gli1-2 repetitive domain is arranged by codons and suggested repeats are arrayed vertically. A consensus structure is given below. The vertical line separates the conserved first three codons of each repeat motif from the variable-length glutamine-rich part of the repeat