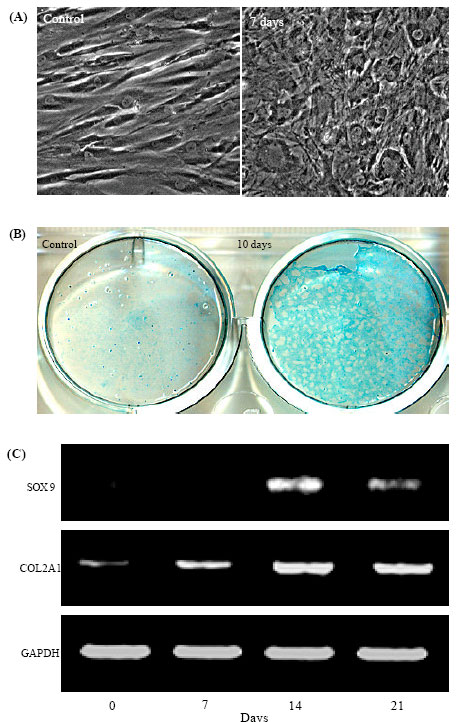
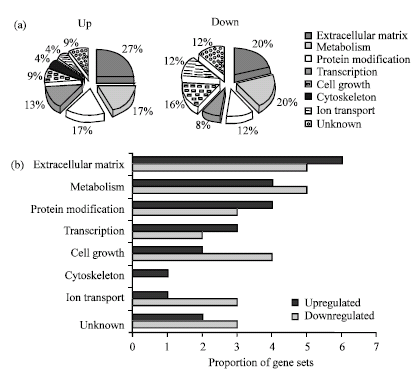
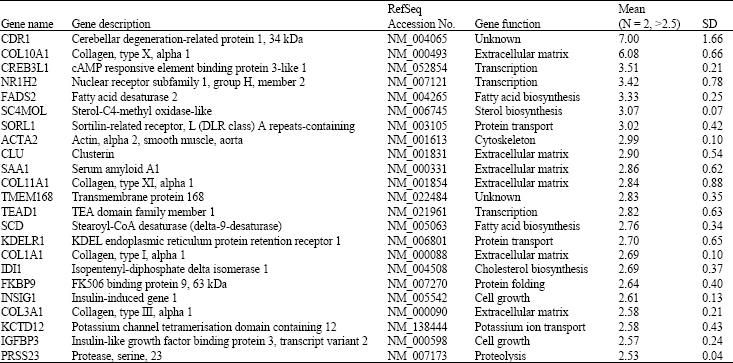
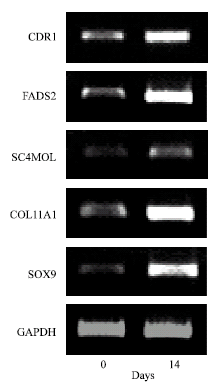
Research Article
Gene Expression Changes During the Chondrogenic Differentiation of Human Mesenchymal Stem Cells
Division of Molecular Life Science, School of Medicine, Tokai University, Bohseidai, Isehara, Kanagawa 259-1193, Japan
So Tsukahara
Division of Molecular Life Science, School of Medicine, Tokai University, Bohseidai, Isehara, Kanagawa 259-1193, Japan
Kenichi Yoshida
Division of Molecular Life Science, School of Medicine, Tokai University, Bohseidai, Isehara, Kanagawa 259-1193, Japan
Ituro Inoue
Division of Molecular Life Science, School of Medicine, Tokai University, Bohseidai, Isehara, Kanagawa 259-1193, Japan