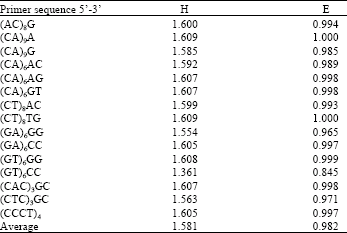
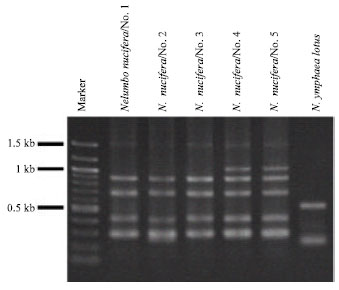
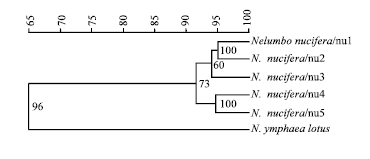
Research Article
Genetic Relationships in a Population of Nelumbo nucifera Gaertn. (Nelumbonaceae)
Department of Biology, Faculty of Science, Khan Kaen University, Khan Kaen 40002, Thailand
Runglawan Sudmoon
Department of Biochemistry, Faculty of Science, Khan Kaen University, Khan Kaen 40002, Thailand
Tawatchai Tanee
Faculty of Environment and Resource Studies, Mahasarakham University, Mahasarakham 44000, Thailand
Piya Mokkamul
Department of Biochemistry, Faculty of Science, Khan Kaen University, Khan Kaen 40002, Thailand
Alongkoad Tanomtong
Department of Biochemistry, Faculty of Science, Khan Kaen University, Khan Kaen 40002, Thailand