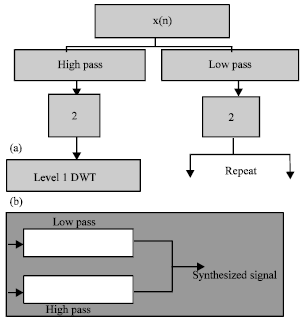
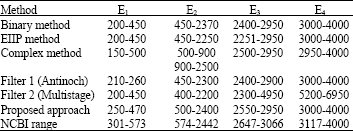
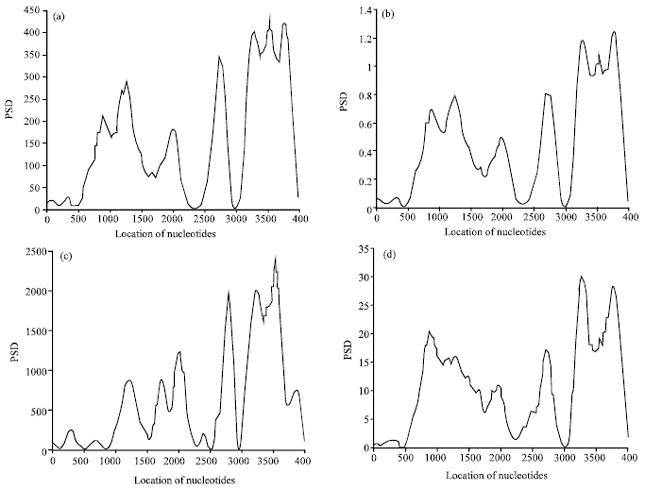
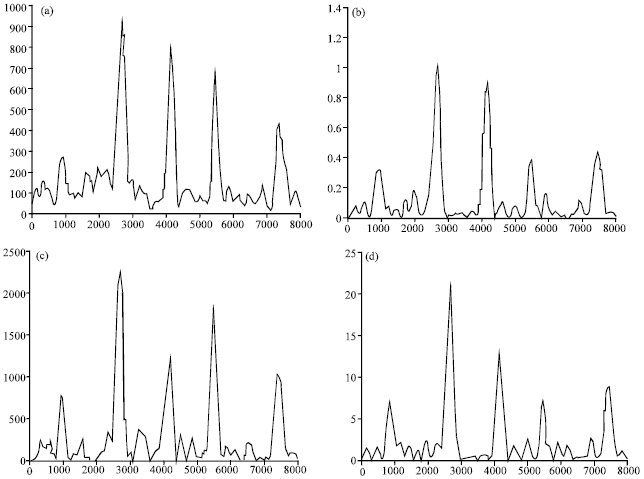
Research Article
A Novel Optimized Approach for Gene Identification in DNA Sequences
College of Computer and Information Sciences, King Faisal University, P.O. Box 55028, Al-Hassa 31982, Saudi Arabia
Azween Abdullah
Department of IT, University Technology PETRONAS, Malaysia
Khalid Buragga
College of Computer and Information Sciences, King Faisal University, P.O. Box 55028, Al-Hassa 31982, Saudi Arabia