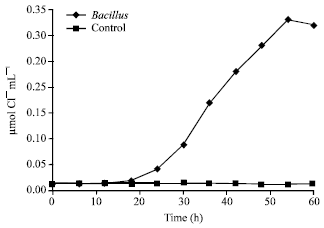
Research Article
Biodegradation of Low Concentration of Monochloroacetic Acid-Degrading Bacillus sp. TW1 Isolated from Terengganu Water Treatment and Distribution Plant
Department of Industrial Biotechnology, Faculty of Biosciences and Bioengineering, Universiti Teknologi Malaysia, 81300 Skudai, Johor, Malaysia
F. Huyop
Department of Industrial Biotechnology, Faculty of Biosciences and Bioengineering, Universiti Teknologi Malaysia, 81300 Skudai, Johor, Malaysia
A.H. Zulkifly
Department of Biotechnology, Faculty of Agriculture and Biotechnology, University Sultan Zainal Abidin, 20400 Kuala Terengganu, Terengganu, Malaysia
D.D. Roslan
Department of Industrial Biotechnology, Faculty of Biosciences and Bioengineering, Universiti Teknologi Malaysia, 81300 Skudai, Johor, Malaysia
A.A.A. Hamid
Department of Industrial Biotechnology, Faculty of Biosciences and Bioengineering, Universiti Teknologi Malaysia, 81300 Skudai, Johor, Malaysia
S. Hamdan
Department of Industrial Biotechnology, Faculty of Biosciences and Bioengineering, Universiti Teknologi Malaysia, 81300 Skudai, Johor, Malaysia
F. Huyop
Department of Industrial Biotechnology, Faculty of Biosciences and Bioengineering, Universiti Teknologi Malaysia, 81300 Skudai, Johor, Malaysia