Research Article
Image Processing for Synthesizing Lung Nodules: A Experimental Study
Department of Electronic and Communications Engineering, Delhi College of Engineering, New Delhi, India
G. Senthil Kumar
Department of Electronic and Communications Engineering, Delhi College of Engineering, New Delhi, India









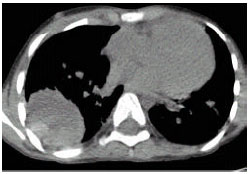
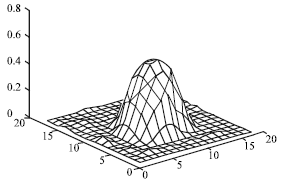
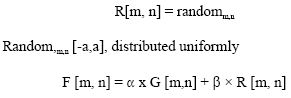
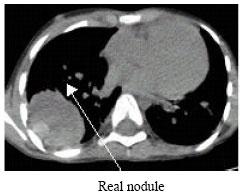
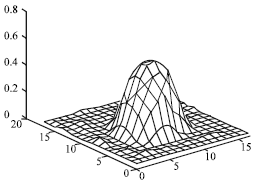
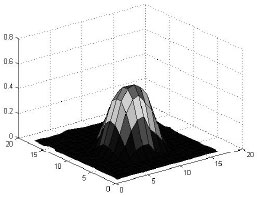
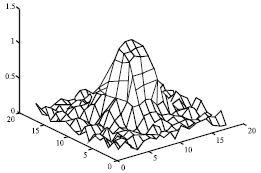
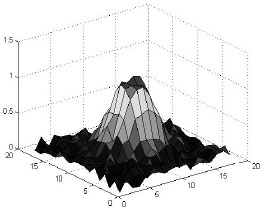
Annapoorani C L Reply
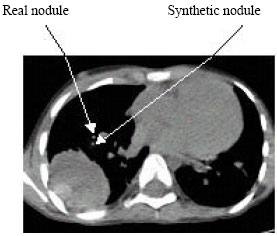
Its an innovative part of creating synthetic lung nodules using MATLAB, that is used for technical computing. But its making me to think why specifically LUNG NODULES are chosen for image processing and not any other nodules...!?