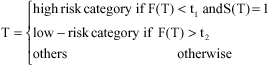Research Article
Analysis of Gene Expression Profile to Select Patient Samples for Outcome Prediction
Department of Computer Science, S.D.N.B. Vaishnav College For Women, Chromepet, Chennai-44, India
S. Jayalakshmi
Department of Computer Science, S.D.N.B. Vaishnav College For Women, Chromepet, Chennai-44, India
S.P. Rajagopalan
School of Computer Science and Engineering, M.G.R. University, Chennai, India