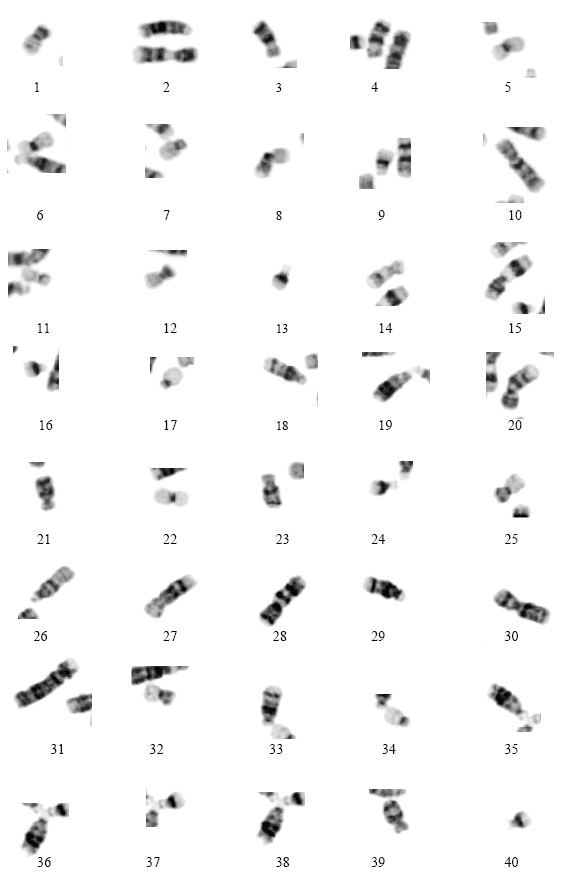
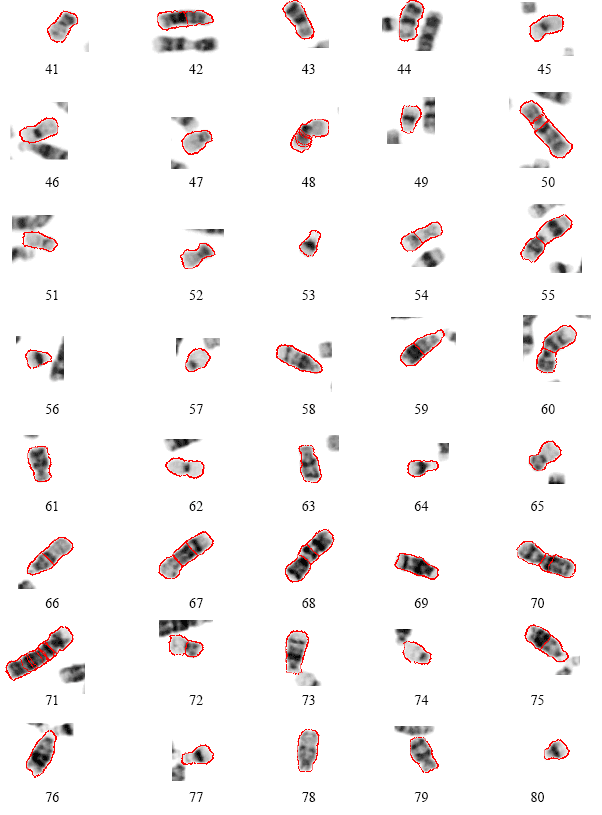
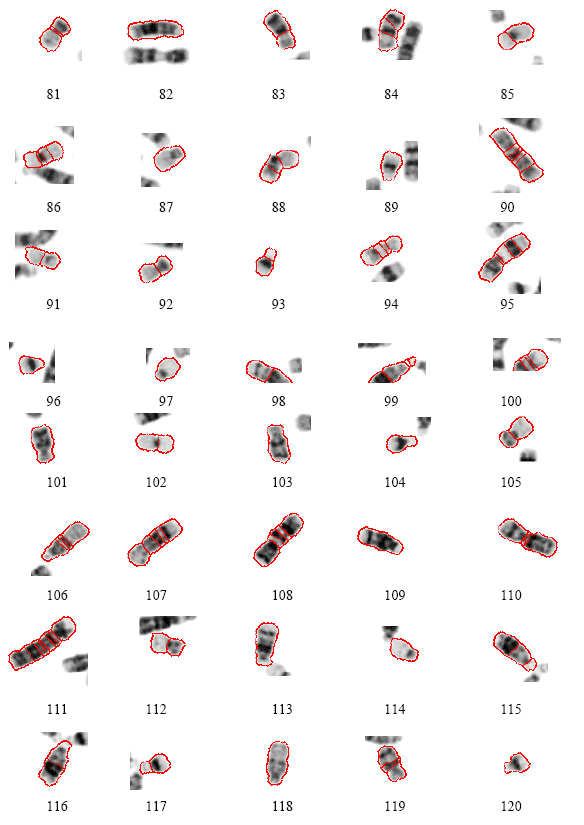
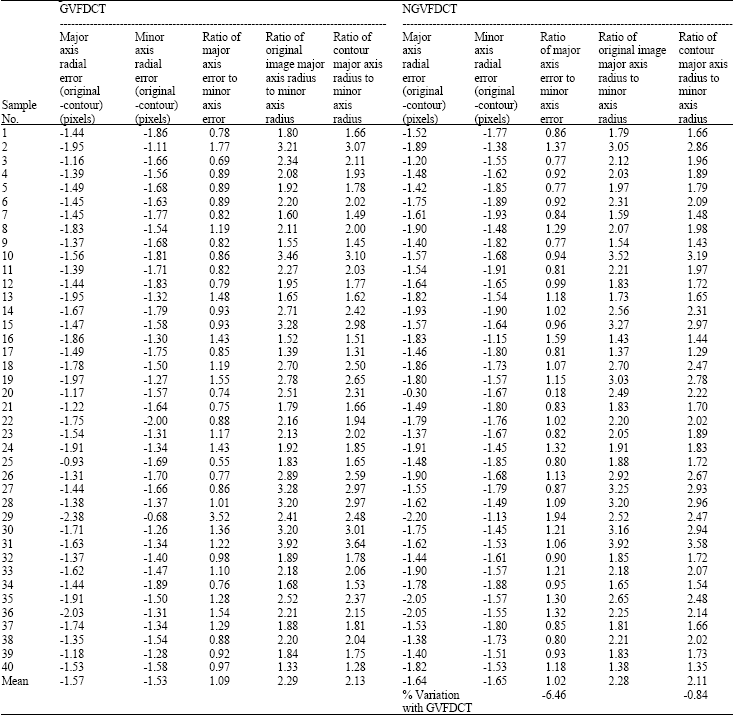
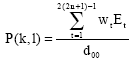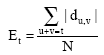Research Article
A Study into the Merit of the DCT Based NGVF Active Contours in Comparison with DCT Based GVF Active Contours
Center for Medical Electronics, Department of Electronics and Communication Engineering, Anna University, Chennai, India
G. Ravindran
Faculty of Information and Communication Engineering, Anna University, Chennai, India