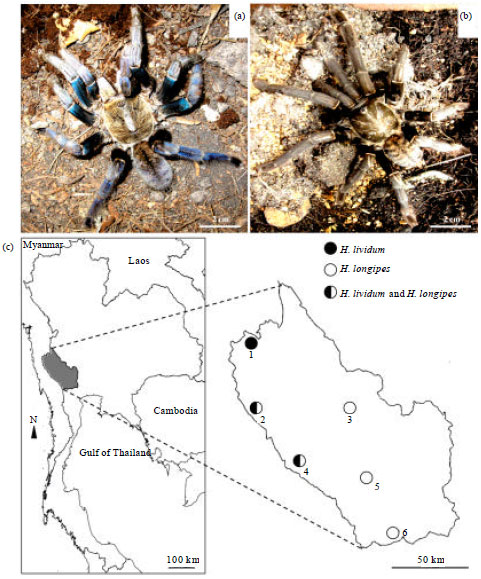
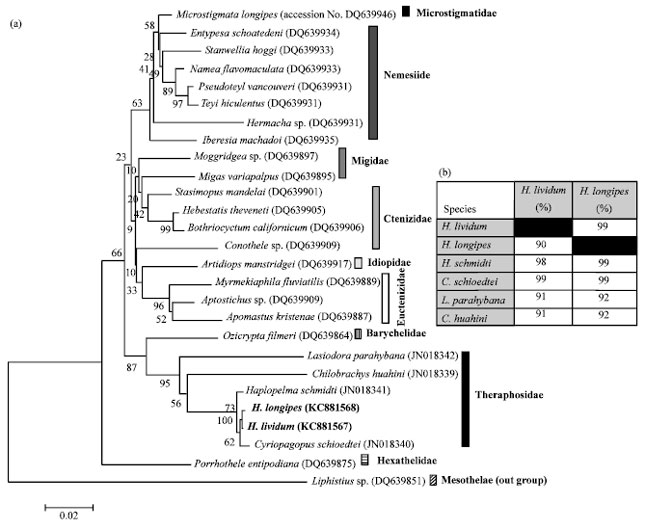
Short Communication
Phylogenetic Relationships of Two Earth Tiger Tarantulas, Haplopelma lividum and H. longipes (Araneae, Theraphosidae), within the Infraorder Mygalomorph Using 28S Ribosomal DNA Sequences
Faculty of Environmental Culture and Ecotourism, Srinakharinwirot University, Bangkok, 10110, Thailand
Manaporn Manaboon
Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, 50200, Thailand
Busaba Panyarachun
Department of Anatomy, Faculty of Medicine, Srinakharinwirot University, Bangkok, 10110, Thailand
Udomsri Showpittapornchai
Department of Anatomy, Faculty of Medicine, Srinakharinwirot University, Bangkok, 10110, Thailand