Research Article
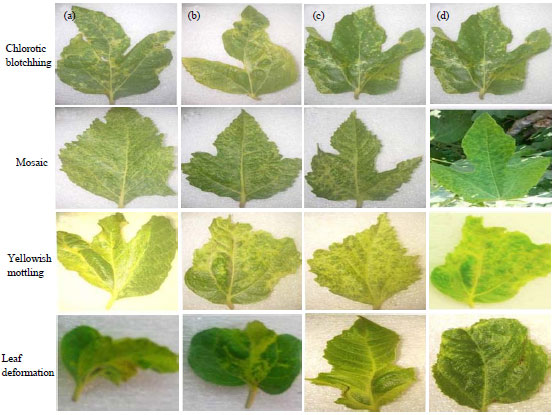
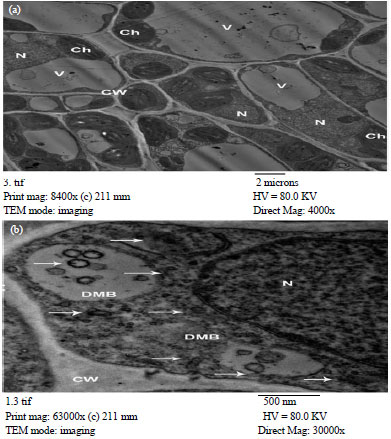
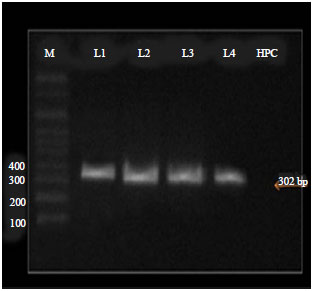
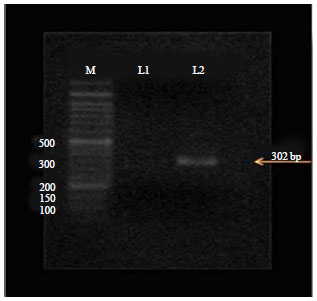
Partial Nucleotide Sequence of the Family Bunyaviridae Associated with a Mosaic-Diseased Fig. in North Egypt
Department of Agriculture Botany, Faculty of Agriculture, Suez Canal University, Ismailia, Egypt
Azza G. Farag
Department of Virus and Phytoplasma Research, Plant Pathology Research Institute, Agriculture Research Center, Giza, Egypt Department of Biotechnology, Faculty of Science, Taif University, Kingdom of Saudi Arabia
Essam K. Elbeshehy
Department of Agriculture Botany, Faculty of Agriculture, Suez Canal University, Ismailia, Egypt Department of Biology, Faculty of Science, Jeddah University, Kingdom of Saudi Arabia
Ahmed K. El Attar
Department of Virus and Phytoplasma Research, Plant Pathology Research Institute, Agriculture Research Center, Giza, Egypt
Sherif A. Sabik
Department of Virus and Phytoplasma Research, Plant Pathology Research Institute, Agriculture Research Center, Giza, Egypt