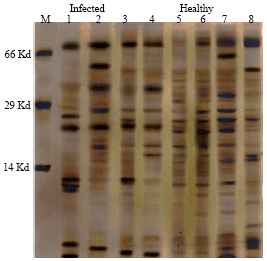
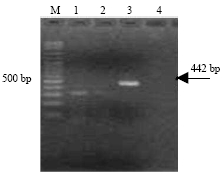
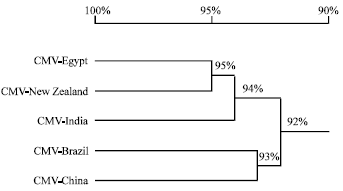
Research Article
Partial Characterization of an Isolate of Cucumber Mosaic Virus from Ismailia Governorate
Department of Agricultural Botany and Plant Pathology, Faculty of Agriculture, Suez Canal University, Egypt
A.A.A. Sallam
Department of Agricultural Botany and Plant Pathology, Faculty of Agriculture, Suez Canal University, Egypt