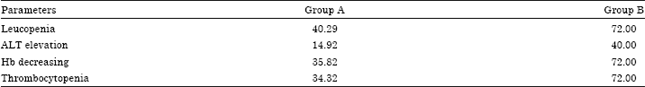
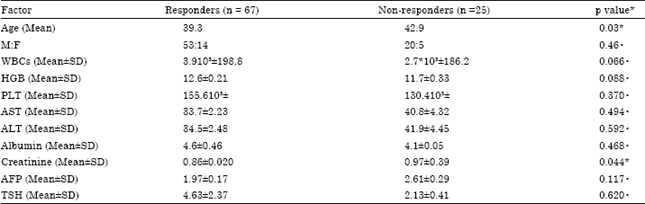
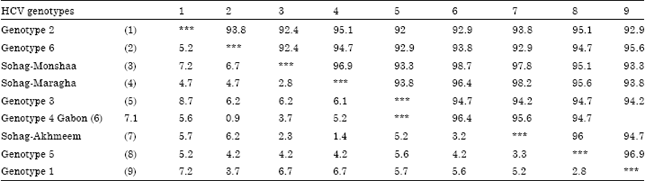
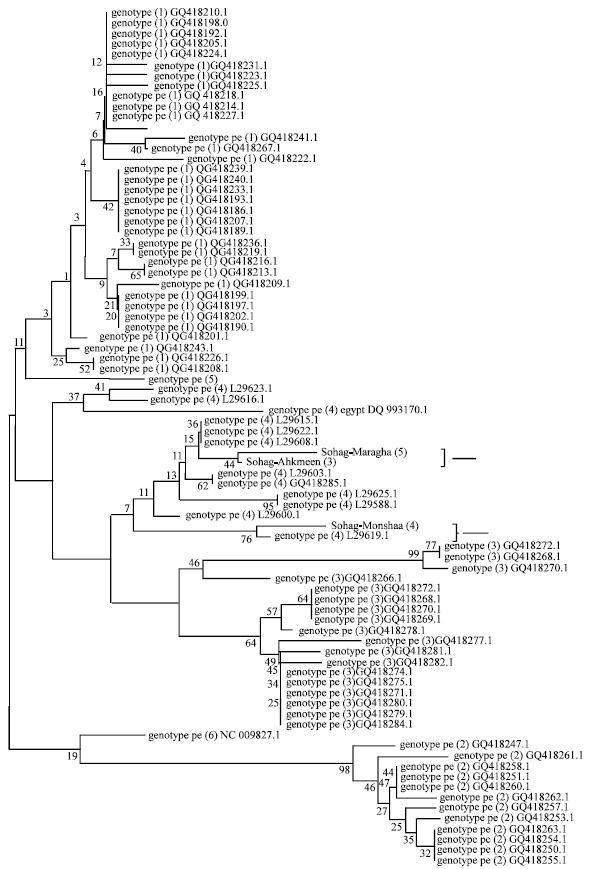
Research Article
Genetic Variations in a Conserved 5'-untranslated Region of Hepatitis C Virus Isolated from Egypt
Genetic Engineering and Biotechnology Research Institute (GEBRI), Sadat City Branch, Minufiya University, Minufiya, Egypt
M. Osman
Genetic Engineering and Biotechnology Research Institute (GEBRI), Sadat City Branch, Minufiya University, Minufiya, Egypt
M. El-Shahat
Genetic Engineering and Biotechnology Research Institute (GEBRI), Sadat City Branch, Minufiya University, Minufiya, Egypt
Medhat H. Hashem
Genetic Engineering and Biotechnology Research Institute (GEBRI), Sadat City Branch, Minufiya University, Minufiya, Egypt
Amal Mahmoud
Genetic Engineering and Biotechnology Research Institute (GEBRI), Sadat City Branch, Minufiya University, Minufiya, Egypt
Hosni Dahi
Genetic Engineering and Biotechnology Research Institute (GEBRI), Sadat City Branch, Minufiya University, Minufiya, Egypt