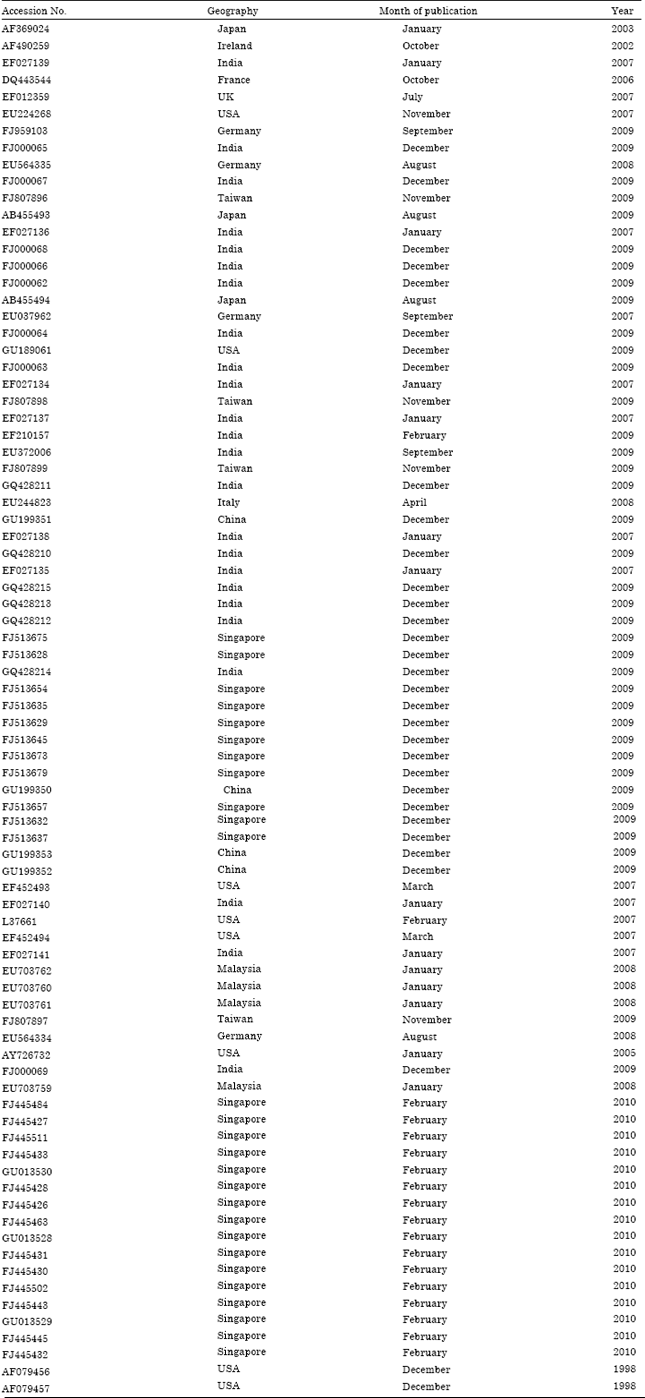
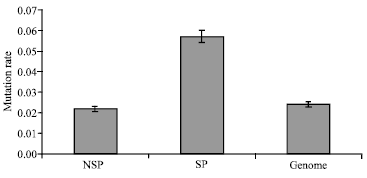
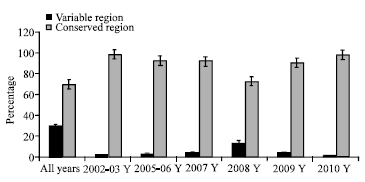
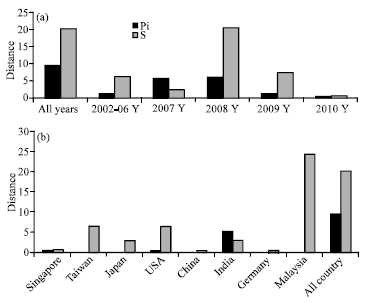
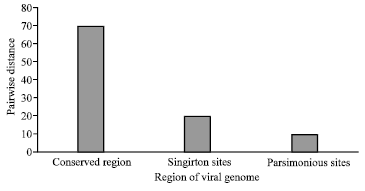
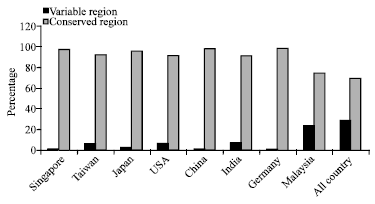
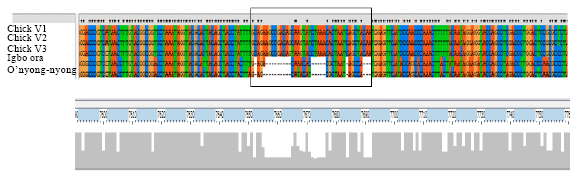
Research Article
Mutational Patterns and DNA Barcode for Diagnosing Chikungunya Virus
Centre for Advanced Study in Marine Biology, Faculty of Marine Biology, Annamalai University, Parangipettai, Tamil Nadu, India
B.A. John
Department of Biotechnology, Kulliyyah of Science, International Islamic University Malaysia, Jalan Sultan Ahmad Shah, Bandar Indera Mahkota, 25200, Kuantan Pahang, Malaysia
S.A. Khan
Centre for Advanced Study in Marine Biology, Faculty of Marine Biology, Annamalai University, Parangipettai, Tamil Nadu, India
P.S. Lyla
Centre for Advanced Study in Marine Biology, Faculty of Marine Biology, Annamalai University, Parangipettai, Tamil Nadu, India
A.S. Kamaleson
Division of Biological Oceanography, National Institute of Oceanography, Dona Paula, Goa, India