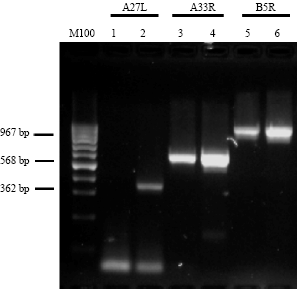
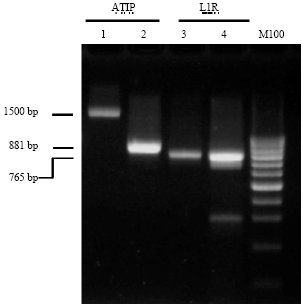
Research Article
Molecular Characterization of Enzootic Camelpox Virus in the Eastern Kingdom of Saudi Arabia
Central Biotechnology Laboratory, College of Veterinary Medicine and Animal Resources, Bldg. 999, King Faisal University, Saudi Arabia
A. Abdelmohsen Al-Naeem
Department of Clinical Studies, Epidemiology Section, College of Veterinary Medicine and Animal Resources, Bldg. 999, King Faisal University, Saudi Arabia